Workflow Type: Galaxy
Frozen
Frozen
Stable
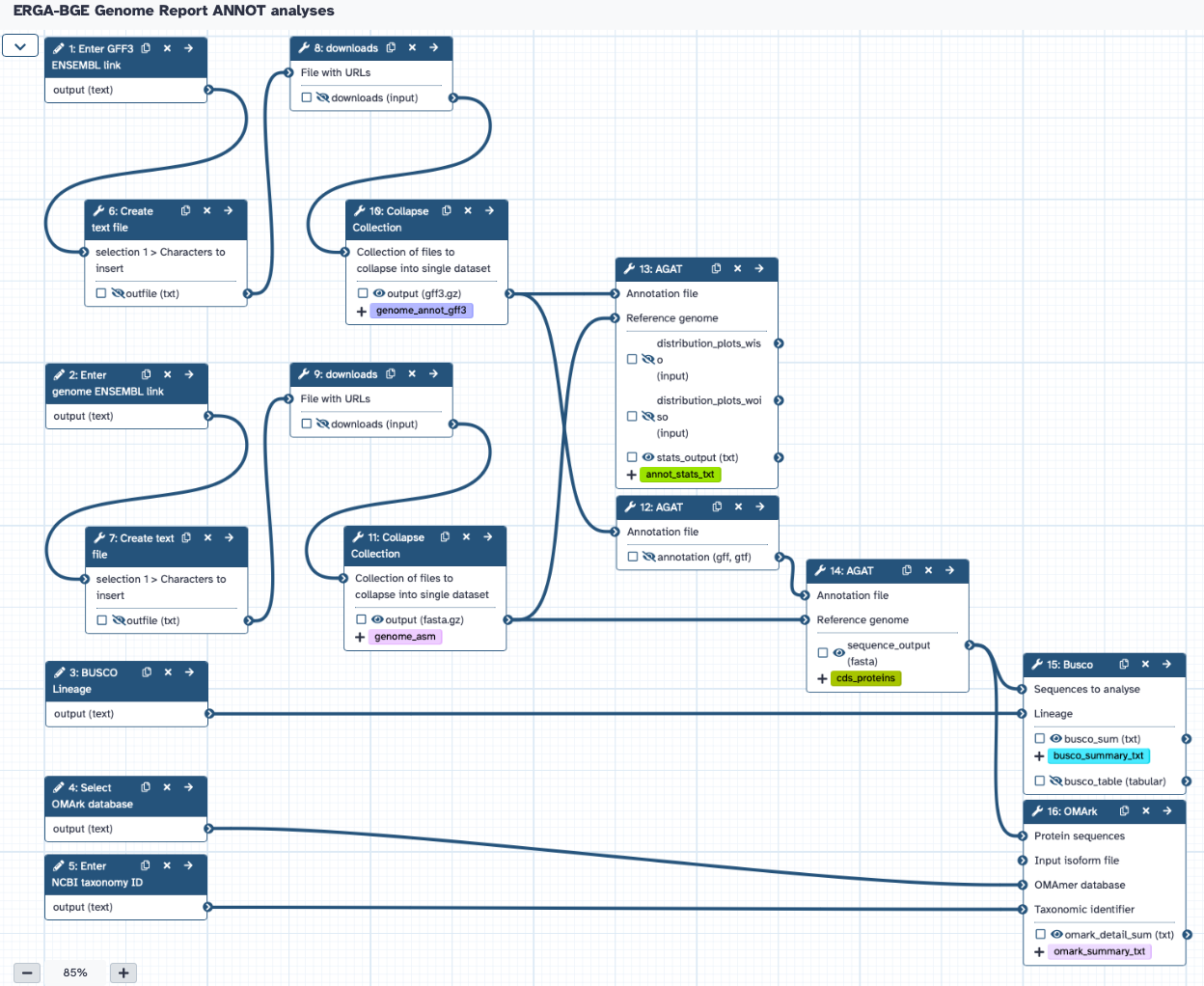
The workflow requires the user to provide:
- ENSEMBL link address of the annotation GFF3 file
- ENSEMBL link address of the assembly FASTA file
- NCBI taxonomy ID
- BUSCO lineage
- OMArk database
Thw workflow will produce statistics of the annotation based on AGAT, BUSCO and OMArk.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| BUSCO Lineage | BUSCO Lineage | Choose the (eukaryotic) BUSCO lineage that corresponds to the assembled species. EXAMPLE: mammalia_odb10 |
|
| Enter GFF3 ENSEMBL link | Enter GFF3 ENSEMBL link | EXAMPLE: https://ftp.ebi.ac.uk/pub/ensemblorganisms/Arca_noae/GCA_964261245.1/ensembl/geneset/2024_11/genes.gff3 OR https://ftp.ensembl.org/pub/rapid-release/species/Apodemus_agrarius/GCA_964023405.1/ensembl/geneset/2024_05/Apodemus_agrarius-GCA_964023405.1-2024_05-genes.gff3.gz |
|
| Enter NCBI taxonomy ID | Enter NCBI taxonomy ID | EXAMPLE: 39030 (Can be obtained from: https://www.ncbi.nlm.nih.gov/taxonomy) |
|
| Enter genome ENSEMBL link | Enter genome ENSEMBL link | EXAMPLE: https://ftp.ebi.ac.uk/pub/ensemblorganisms/Arca_noae/GCA_964261245.1/genome/unmasked.fa.gz OR https://ftp.ensembl.org/pub/rapid-release/species/Apodemus_agrarius/GCA_964023405.1/ensembl/genome/Apodemus_agrarius-GCA_964023405.1-unmasked.fa.gz |
|
| Is the GFF compressed? (gff3.gz) | Is the GFF compressed? (gff3.gz) | n/a |
|
| Select OMArk database | Select OMArk database | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 6 | Create text file | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_text_file_with_recurring_lines/9.3+galaxy1 |
| 7 | Create text file | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_text_file_with_recurring_lines/9.3+galaxy1 |
| 8 | downloads | lftp |
| 9 | downloads | lftp |
| 10 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 11 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 12 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 13 | Pick parameter value | toolshed.g2.bx.psu.edu/repos/iuc/pick_value/pick_value/0.2.0 |
| 14 | AGAT | toolshed.g2.bx.psu.edu/repos/bgruening/agat/agat/1.2.0+galaxy1 |
| 15 | AGAT | toolshed.g2.bx.psu.edu/repos/bgruening/agat/agat/1.2.0+galaxy2 |
| 16 | AGAT | toolshed.g2.bx.psu.edu/repos/bgruening/agat/agat/1.2.0+galaxy1 |
| 17 | Busco | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.5.0+galaxy0 |
| 18 | OMArk | toolshed.g2.bx.psu.edu/repos/iuc/omark/omark/0.3.0+galaxy2 |
Version History
Version 2 (latest) Created 5th Feb 2025 at 21:21 by Diego De Panis
No revision comments
Frozen
 Version-2
Version-235eb999
Version 1 (earliest) Created 9th Aug 2024 at 15:14 by Diego De Panis
Initial commit
Frozen
 Version-1
Version-1c0ef3ff
 Creators and Submitter
Creators and SubmitterCreator
Additional credit
ERGA
Submitter
Discussion Channel
License
Activity
Views: 2867 Downloads: 427 Runs: 43
Created: 9th Aug 2024 at 15:14
Last updated: 24th Feb 2025 at 15:24
Annotated Properties
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections- Galaxy-Workflow-ERGA-BGE_Genome_Report_ANNOT_analyses.gaRemoteMain Workflow
- annot_rev.pngDiagramRemote
 View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-3679-9585
https://orcid.org/0000-0002-3679-9585