Workflow Type: Galaxy
Open
Frozen
Stable
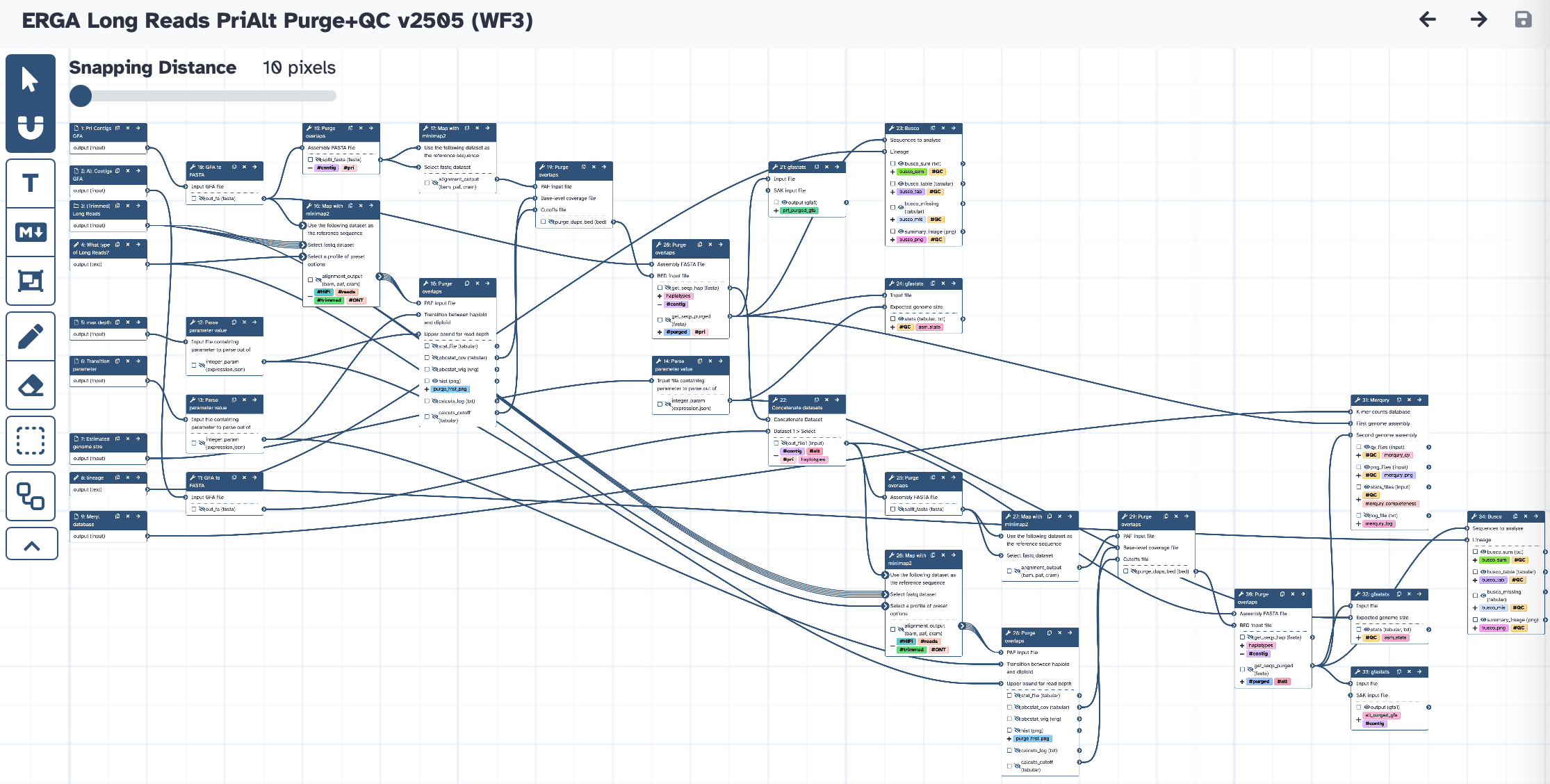
The workflow takes a Long Reads collection, Pri/Alt contigs, and the values for transition parameter and max coverage depth (calculated from WF1) to run Purge_Dups. It produces purged Pri and Alt contigs assemblies, and runs all the QC analysis (gfastats, BUSCO, and Merqury).
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| (Trimmed) Long Reads | (Trimmed) Long Reads | Collection of Long reads in fastq format |
|
| Alt Contigs GFA | Alt Contigs GFA | Alternate contigs in GFA format from the Hifiasm run results: alt_contigs_gfa |
|
| Estimated genome size | Estimated genome size | Select the est_genome_size result obtained during profiling |
|
| Meryl database | Meryl database | Select the meryl_db result obtained during profiling |
|
| Pri Contigs GFA | Pri Contigs GFA | Primary contigs in GFA format from the Hifiasm run results: pri_contigs_gfa |
|
| Transition parameter | Transition parameter | Select the transition_param result obtained during profiling |
|
| What type of Long Reads? | What type of Long Reads? | Select map-hifi if the long reads are PacBio HiFi (default), map-ont if the long reads are ONT |
|
| lineage | lineage | lineage for BUSCO, e.g.: arthropoda_odb10, vertebrata_odb10, mammalia_odb10, aves_odb10, tetrapoda_odb10 ... |
|
| max depth | max depth | Select the max_depth result obtained during profiling |
|
Steps
| ID | Name | Description |
|---|---|---|
| 9 | GFA to FASTA | toolshed.g2.bx.psu.edu/repos/iuc/gfa_to_fa/gfa_to_fa/0.1.2 |
| 10 | GFA to FASTA | toolshed.g2.bx.psu.edu/repos/iuc/gfa_to_fa/gfa_to_fa/0.1.2 |
| 11 | Parse parameter value | param_value_from_file |
| 12 | Parse parameter value | param_value_from_file |
| 13 | Parse parameter value | param_value_from_file |
| 14 | Purge overlaps | toolshed.g2.bx.psu.edu/repos/iuc/purge_dups/purge_dups/1.2.6+galaxy0 |
| 15 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.28+galaxy1 |
| 16 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.28+galaxy1 |
| 17 | Purge overlaps | toolshed.g2.bx.psu.edu/repos/iuc/purge_dups/purge_dups/1.2.6+galaxy0 |
| 18 | Purge overlaps | toolshed.g2.bx.psu.edu/repos/iuc/purge_dups/purge_dups/1.2.6+galaxy0 |
| 19 | Purge overlaps | toolshed.g2.bx.psu.edu/repos/iuc/purge_dups/purge_dups/1.2.6+galaxy0 |
| 20 | gfastats | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.11+galaxy0 |
| 21 | Concatenate datasets | cat1 |
| 22 | Busco | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.8.0+galaxy1 |
| 23 | gfastats | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.11+galaxy0 |
| 24 | Purge overlaps | toolshed.g2.bx.psu.edu/repos/iuc/purge_dups/purge_dups/1.2.6+galaxy0 |
| 25 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.28+galaxy1 |
| 26 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.28+galaxy1 |
| 27 | Purge overlaps | toolshed.g2.bx.psu.edu/repos/iuc/purge_dups/purge_dups/1.2.6+galaxy0 |
| 28 | Purge overlaps | toolshed.g2.bx.psu.edu/repos/iuc/purge_dups/purge_dups/1.2.6+galaxy0 |
| 29 | Purge overlaps | toolshed.g2.bx.psu.edu/repos/iuc/purge_dups/purge_dups/1.2.6+galaxy0 |
| 30 | Merqury | toolshed.g2.bx.psu.edu/repos/iuc/merqury/merqury/1.3+galaxy4 |
| 31 | gfastats | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.11+galaxy0 |
| 32 | gfastats | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.11+galaxy0 |
| 33 | Busco | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.8.0+galaxy1 |
Version History
Version 2 (latest) Created 1st Jun 2025 at 12:24 by Diego De Panis
No revision comments
Open
 master
master8ad8be8
Version 1 (earliest) Created 24th Sep 2024 at 22:44 by Diego De Panis
Initial commit
Frozen
 Version-1
Version-1b1bb0ff
 Creators and Submitter
Creators and SubmitterCreator
Additional credit
ERGA
Submitter
Discussion Channel
Tools
License
Activity
Views: 2360 Downloads: 330 Runs: 53
Created: 24th Sep 2024 at 22:44
Last updated: 1st Jun 2025 at 12:25
Annotated Properties
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-3679-9585
https://orcid.org/0000-0002-3679-9585