Workflow Type: Galaxy
Open
Frozen
Microbiome - QC and Contamination Filtering
Associated Tutorial
This workflows is part of the tutorial Nanopore Preprocessing, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Tutorial Author(s): Bérénice Batut, Engy Nasr, Paul Zierep
Tutorial Contributor(s): Hans-Rudolf Hotz, Wolfgang Maier
Workflow Author(s): Bérénice Batut, Engy Nasr, Paul Zierep
Funder(s): Gallantries: Bridging Training Communities in Life Science, Environment and Health, EOSC-Life
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| collection_of_all_samples | collection_of_all_samples | Nanopore reads of each sample are all in a single fastq or fastq.gz file |
|
| samples_profile | samples_profile | based on the lab preparation of the samples during sequencing, there should be a sample profile better than the other, to be chosen as an optional input to Minimap2. e.g. PacBio/Oxford Nanpore For more details check: https://github.com/lh3/minimap2?tab=readme-ov-file#use-cases |
|
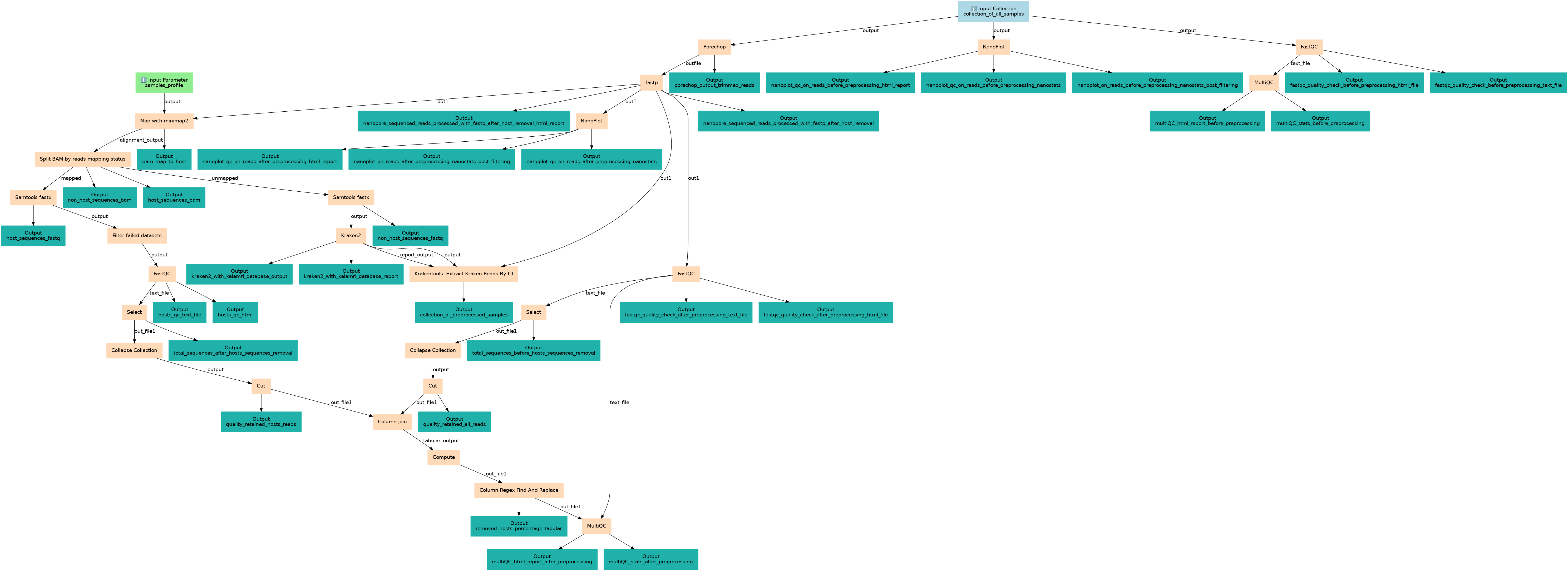
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Porechop | Preprocessing (Trimming) toolshed.g2.bx.psu.edu/repos/iuc/porechop/porechop/0.2.4+galaxy0 |
| 3 | NanoPlot | Quality Check Before Preprocessing toolshed.g2.bx.psu.edu/repos/iuc/nanoplot/nanoplot/1.42.0+galaxy1 |
| 4 | FastQC | Quality Check Before Preprocessing toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 5 | fastp | Preprocessing toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.23.4+galaxy0 |
| 6 | MultiQC | Quality Control Before and After Preprocessing toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 7 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.28+galaxy0 |
| 8 | NanoPlot | Nanoplot Quality Check After Preprocessing toolshed.g2.bx.psu.edu/repos/iuc/nanoplot/nanoplot/1.42.0+galaxy1 |
| 9 | FastQC | Quality Check After Preprocessing toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 10 | Split BAM by reads mapping status | toolshed.g2.bx.psu.edu/repos/iuc/bamtools_split_mapped/bamtools_split_mapped/2.5.2+galaxy2 |
| 11 | Select | Grep1 |
| 12 | Samtools fastx | toolshed.g2.bx.psu.edu/repos/iuc/samtools_fastx/samtools_fastx/1.15.1+galaxy2 |
| 13 | Samtools fastx | toolshed.g2.bx.psu.edu/repos/iuc/samtools_fastx/samtools_fastx/1.15.1+galaxy2 |
| 14 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 15 | Filter failed datasets | __FILTER_FAILED_DATASETS__ |
| 16 | Kraken2 | toolshed.g2.bx.psu.edu/repos/iuc/kraken2/kraken2/2.1.1+galaxy1 |
| 17 | Cut | Cut1 |
| 18 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 19 | Krakentools: Extract Kraken Reads By ID | toolshed.g2.bx.psu.edu/repos/iuc/krakentools_extract_kraken_reads/krakentools_extract_kraken_reads/1.2+galaxy1 |
| 20 | Select | Grep1 |
| 21 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 22 | Cut | Cut1 |
| 23 | Column join | toolshed.g2.bx.psu.edu/repos/iuc/collection_column_join/collection_column_join/0.0.3 |
| 24 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 25 | Column Regex Find And Replace | toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 26 | MultiQC | Quality Control Before and After Preprocessing toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| porechop_output_trimmed_reads | porechop_output_trimmed_reads | n/a |
|
| nanoplot_qc_on_reads_before_preprocessing_nanostats | nanoplot_qc_on_reads_before_preprocessing_nanostats | n/a |
|
| nanoplot_on_reads_before_preprocessing_nanostats_post_filtering | nanoplot_on_reads_before_preprocessing_nanostats_post_filtering | n/a |
|
| nanoplot_qc_on_reads_before_preprocessing_html_report | nanoplot_qc_on_reads_before_preprocessing_html_report | n/a |
|
| fastqc_quality_check_before_preprocessing_html_file | fastqc_quality_check_before_preprocessing_html_file | n/a |
|
| fastqc_quality_check_before_preprocessing_text_file | fastqc_quality_check_before_preprocessing_text_file | n/a |
|
| nanopore_sequenced_reads_processed_with_fastp_after_host_removal | nanopore_sequenced_reads_processed_with_fastp_after_host_removal | n/a |
|
| nanopore_sequenced_reads_processed_with_fastp_after_host_removal_html_report | nanopore_sequenced_reads_processed_with_fastp_after_host_removal_html_report | n/a |
|
| multiQC_stats_before_preprocessing | multiQC_stats_before_preprocessing | n/a |
|
| multiQC_html_report_before_preprocessing | multiQC_html_report_before_preprocessing | n/a |
|
| bam_map_to_host | bam_map_to_host | n/a |
|
| nanoplot_qc_on_reads_after_preprocessing_html_report | nanoplot_qc_on_reads_after_preprocessing_html_report | n/a |
|
| nanoplot_on_reads_after_preprocessing_nanostats_post_filtering | nanoplot_on_reads_after_preprocessing_nanostats_post_filtering | n/a |
|
| nanoplot_qc_on_reads_after_preprocessing_nanostats | nanoplot_qc_on_reads_after_preprocessing_nanostats | n/a |
|
| fastqc_quality_check_after_preprocessing_text_file | fastqc_quality_check_after_preprocessing_text_file | n/a |
|
| fastqc_quality_check_after_preprocessing_html_file | fastqc_quality_check_after_preprocessing_html_file | n/a |
|
| non_host_sequences_bam | non_host_sequences_bam | n/a |
|
| host_sequences_bam | host_sequences_bam | n/a |
|
| total_sequences_before_hosts_sequences_removal | total_sequences_before_hosts_sequences_removal | n/a |
|
| host_sequences_fastq | host_sequences_fastq | n/a |
|
| non_host_sequences_fastq | non_host_sequences_fastq | n/a |
|
| kraken2_with_kalamri_database_output | kraken2_with_kalamri_database_output | n/a |
|
| kraken2_with_kalamri_database_report | kraken2_with_kalamri_database_report | n/a |
|
| quality_retained_all_reads | quality_retained_all_reads | n/a |
|
| hosts_qc_text_file | hosts_qc_text_file | n/a |
|
| hosts_qc_html | hosts_qc_html | n/a |
|
| collection_of_preprocessed_samples | collection_of_preprocessed_samples | n/a |
|
| total_sequences_after_hosts_sequences_removal | total_sequences_after_hosts_sequences_removal | n/a |
|
| quality_retained_hosts_reads | quality_retained_hosts_reads | n/a |
|
| removed_hosts_percentage_tabular | removed_hosts_percentage_tabular | n/a |
|
| multiQC_html_report_after_preprocessing | multiQC_html_report_after_preprocessing | n/a |
|
| multiQC_stats_after_preprocessing | multiQC_stats_after_preprocessing | n/a |
|
Version History
3.0 (latest) Created 16th Jul 2024 at 14:05 by Helena Rasche
Added/updated 4 files
Open
 master
master0a8feb6
5.0 (earliest) Created 25th Jun 2024 at 10:56 by Helena Rasche
Added/updated 4 files
Frozen
 5.0
5.071b4f35
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 552 Downloads: 186
Created: 25th Jun 2024 at 10:55
Last updated: 25th Jun 2024 at 10:56
 Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992