Workflow Type: Galaxy
Open
Frozen
Building an amplicon sequence variant (ASV) table from 16S data using DADA2
Associated Tutorial
This workflows is part of the tutorial Building an amplicon sequence variant (ASV) table from 16S data using DADA2, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Tutorial Author(s): Bérénice Batut
Tutorial Contributor(s): Matthias Bernt, Clea Siguret
Workflow Author(s): , Bérénice Batut
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Raw reads | Raw reads | raw_reads |
|
| header | header | header |
|
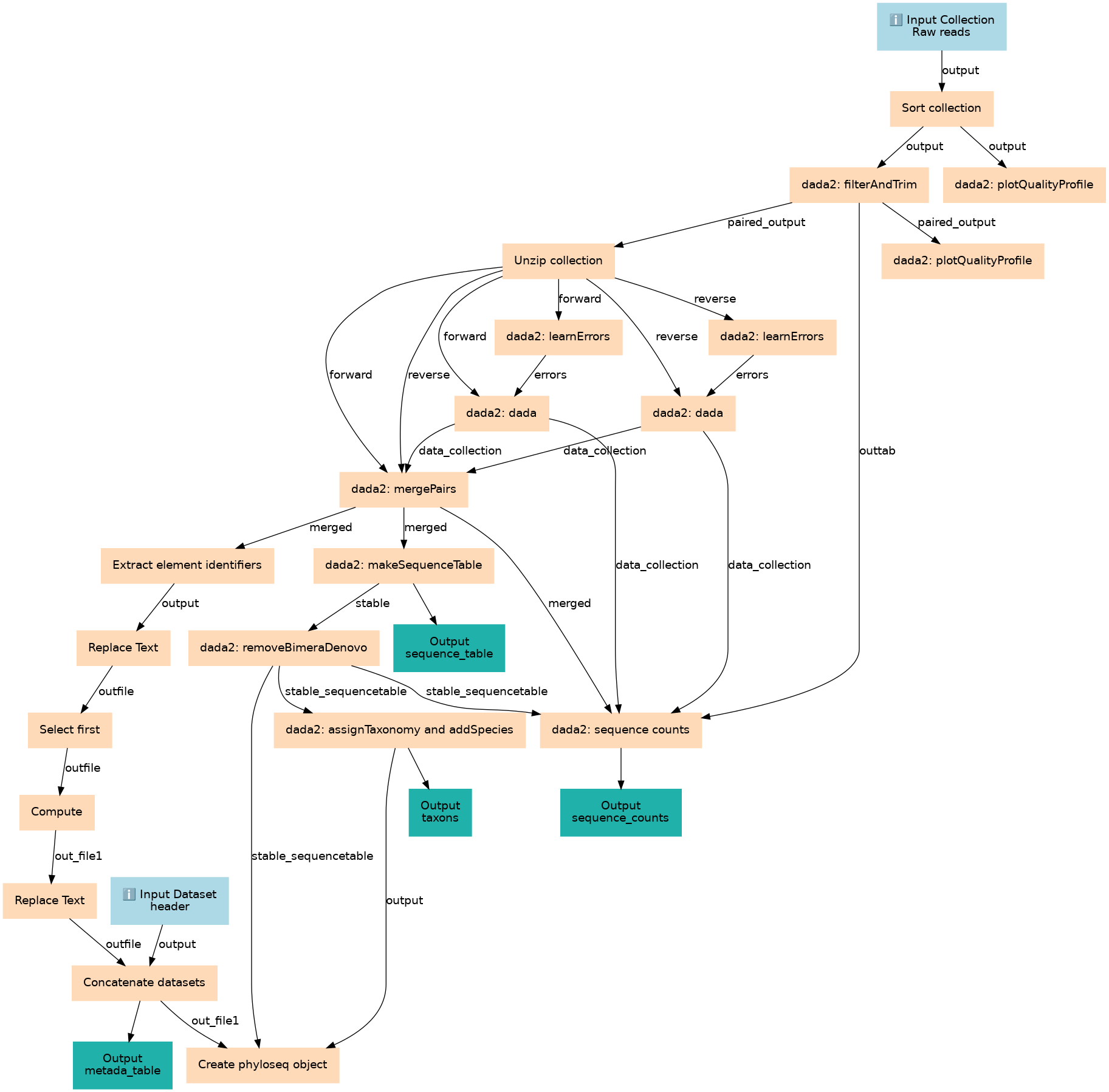
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Sort collection | __SORTLIST__ |
| 3 | dada2: filterAndTrim | toolshed.g2.bx.psu.edu/repos/iuc/dada2_filterandtrim/dada2_filterAndTrim/1.30.0+galaxy0 |
| 4 | dada2: plotQualityProfile | toolshed.g2.bx.psu.edu/repos/iuc/dada2_plotqualityprofile/dada2_plotQualityProfile/1.30.0+galaxy0 |
| 5 | Unzip collection | __UNZIP_COLLECTION__ |
| 6 | dada2: plotQualityProfile | toolshed.g2.bx.psu.edu/repos/iuc/dada2_plotqualityprofile/dada2_plotQualityProfile/1.30.0+galaxy0 |
| 7 | dada2: learnErrors | toolshed.g2.bx.psu.edu/repos/iuc/dada2_learnerrors/dada2_learnErrors/1.30.0+galaxy0 |
| 8 | dada2: learnErrors | toolshed.g2.bx.psu.edu/repos/iuc/dada2_learnerrors/dada2_learnErrors/1.30.0+galaxy0 |
| 9 | dada2: dada | toolshed.g2.bx.psu.edu/repos/iuc/dada2_dada/dada2_dada/1.30.0+galaxy0 |
| 10 | dada2: dada | toolshed.g2.bx.psu.edu/repos/iuc/dada2_dada/dada2_dada/1.30.0+galaxy0 |
| 11 | dada2: mergePairs | toolshed.g2.bx.psu.edu/repos/iuc/dada2_mergepairs/dada2_mergePairs/1.30.0+galaxy0 |
| 12 | Extract element identifiers | toolshed.g2.bx.psu.edu/repos/iuc/collection_element_identifiers/collection_element_identifiers/0.0.2 |
| 13 | dada2: makeSequenceTable | toolshed.g2.bx.psu.edu/repos/iuc/dada2_makesequencetable/dada2_makeSequenceTable/1.30.0+galaxy0 |
| 14 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_line/9.3+galaxy1 |
| 15 | dada2: removeBimeraDenovo | toolshed.g2.bx.psu.edu/repos/iuc/dada2_removebimeradenovo/dada2_removeBimeraDenovo/1.30.0+galaxy0 |
| 16 | Select first | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_head_tool/9.3+galaxy1 |
| 17 | dada2: sequence counts | toolshed.g2.bx.psu.edu/repos/iuc/dada2_seqcounts/dada2_seqCounts/1.30.0+galaxy0 |
| 18 | dada2: assignTaxonomy and addSpecies | toolshed.g2.bx.psu.edu/repos/iuc/dada2_assigntaxonomyaddspecies/dada2_assignTaxonomyAddspecies/1.30.0+galaxy0 |
| 19 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 20 | Replace Text | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1 |
| 21 | Concatenate datasets | cat1 |
| 22 | Create phyloseq object | toolshed.g2.bx.psu.edu/repos/iuc/phyloseq_from_dada2/phyloseq_from_dada2/1.46.0+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| sequence_table | sequence_table | n/a |
|
| sequence_counts | sequence_counts | n/a |
|
| taxons | taxons | n/a |
|
| metada_table | metada_table | n/a |
|
Version History
1.0 (latest) Created 16th Jul 2024 at 14:05 by Helena Rasche
Added/updated 4 files
Open
 master
master242aa01
2.0 (earliest) Created 25th Jun 2024 at 10:58 by Helena Rasche
Added/updated 4 files
Frozen
 2.0
2.0411b7a5
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 656 Downloads: 171
Created: 25th Jun 2024 at 10:58
Last updated: 25th Jun 2024 at 10:58
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992