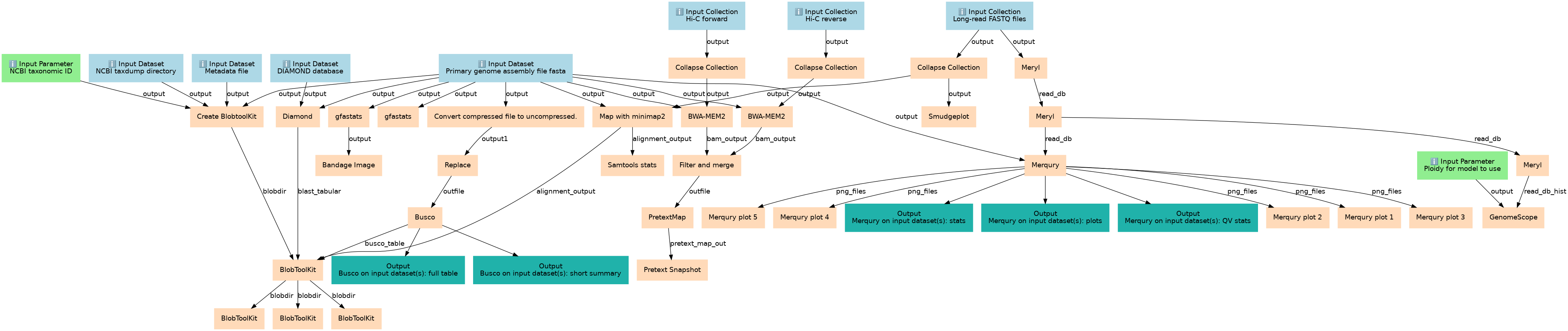
Workflow Type: Galaxy
Open
Frozen
ERGA post-assembly QC
Associated Tutorial
This workflows is part of the tutorial ERGA post-assembly QC, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Cristóbal Gallardo Alba
Tutorial Author(s): Fabian Recktenwald, Cristóbal Gallardo, Tom Brown
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| DIAMOND database | DIAMOND database | n/a |
|
| Hi-C forward | Hi-C forward | n/a |
|
| Hi-C reverse | Hi-C reverse | n/a |
|
| Long-read FASTQ files | Long-read FASTQ files | n/a |
|
| Metadata file | Metadata file | n/a |
|
| NCBI taxdump directory | NCBI taxdump directory | n/a |
|
| NCBI taxonomic ID | NCBI taxonomic ID | n/a |
|
| Ploidy for model to use | Ploidy for model to use | n/a |
|
| Primary genome assembly file (fasta) | Primary genome assembly file (fasta) | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 9 | Meryl | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 10 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 11 | Create BlobtoolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 12 | gfastats | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.2.0+galaxy0 |
| 13 | Convert compressed file to uncompressed. | CONVERTER_gz_to_uncompressed |
| 14 | gfastats | toolshed.g2.bx.psu.edu/repos/bgruening/gfastats/gfastats/1.3.6+galaxy0 |
| 15 | Diamond | toolshed.g2.bx.psu.edu/repos/bgruening/diamond/bg_diamond/2.0.15+galaxy0 |
| 16 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 17 | Collapse Collection | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 18 | Meryl | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 19 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.28+galaxy0 |
| 20 | Smudgeplot | toolshed.g2.bx.psu.edu/repos/galaxy-australia/smudgeplot/smudgeplot/0.2.5+galaxy+2 |
| 21 | Replace | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_find_and_replace/1.1.4 |
| 22 | Bandage Image | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/2022.09+galaxy4 |
| 23 | BWA-MEM2 | toolshed.g2.bx.psu.edu/repos/iuc/bwa_mem2/bwa_mem2/2.2.1+galaxy0 |
| 24 | BWA-MEM2 | toolshed.g2.bx.psu.edu/repos/iuc/bwa_mem2/bwa_mem2/2.2.1+galaxy0 |
| 25 | Merqury | toolshed.g2.bx.psu.edu/repos/iuc/merqury/merqury/1.3+galaxy2 |
| 26 | Meryl | toolshed.g2.bx.psu.edu/repos/iuc/meryl/meryl/1.3+galaxy6 |
| 27 | Samtools stats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_stats/samtools_stats/2.0.4 |
| 28 | Busco | toolshed.g2.bx.psu.edu/repos/iuc/busco/busco/5.4.4+galaxy0 |
| 29 | Filter and merge | toolshed.g2.bx.psu.edu/repos/iuc/bellerophon/bellerophon/1.0+galaxy0 |
| 30 | Merqury plot 2 | __EXTRACT_DATASET__ |
| 31 | Merqury plot 1 | __EXTRACT_DATASET__ |
| 32 | Merqury plot 3 | __EXTRACT_DATASET__ |
| 33 | Merqury plot 5 | __EXTRACT_DATASET__ |
| 34 | Merqury plot 4 | __EXTRACT_DATASET__ |
| 35 | GenomeScope | toolshed.g2.bx.psu.edu/repos/iuc/genomescope/genomescope/2.0+galaxy2 |
| 36 | BlobToolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 37 | PretextMap | toolshed.g2.bx.psu.edu/repos/iuc/pretext_map/pretext_map/0.1.9+galaxy0 |
| 38 | BlobToolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 39 | BlobToolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 40 | BlobToolKit | toolshed.g2.bx.psu.edu/repos/bgruening/blobtoolkit/blobtoolkit/4.0.7+galaxy2 |
| 41 | Pretext Snapshot | toolshed.g2.bx.psu.edu/repos/iuc/pretext_snapshot/pretext_snapshot/0.0.3+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
| Merqury on input dataset(s): stats | Merqury on input dataset(s): stats | n/a |
|
| Merqury on input dataset(s): plots | Merqury on input dataset(s): plots | n/a |
|
| Merqury on input dataset(s): QV stats | Merqury on input dataset(s): QV stats | n/a |
|
| _anonymous_output_7 | _anonymous_output_7 | n/a |
|
| Busco on input dataset(s): full table | Busco on input dataset(s): full table | n/a |
|
| Busco on input dataset(s): short summary | Busco on input dataset(s): short summary | n/a |
|
| _anonymous_output_8 | _anonymous_output_8 | n/a |
|
| _anonymous_output_9 | _anonymous_output_9 | n/a |
|
| _anonymous_output_10 | _anonymous_output_10 | n/a |
|
| _anonymous_output_11 | _anonymous_output_11 | n/a |
|
| _anonymous_output_12 | _anonymous_output_12 | n/a |
|
Version History
2.0 (latest) Created 9th Sep 2024 at 14:24 by Helena Rasche
Added/updated 4 files
Open
 master
master1553de1
3.0 (earliest) Created 25th Jun 2024 at 11:00 by Helena Rasche
Added/updated 4 files
Frozen
 3.0
3.01329bf0
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 726 Downloads: 252
Created: 25th Jun 2024 at 11:00
Last updated: 9th Sep 2024 at 14:24
 Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992