Workflow Type: Galaxy
Open
Frozen
Associated Tutorial
This workflows is part of the tutorial Assembly with Flye - upgraded, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Tutorial Author(s): Anna Syme
Workflow Author(s): Anna Syme
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| long reads | long reads | n/a |
|
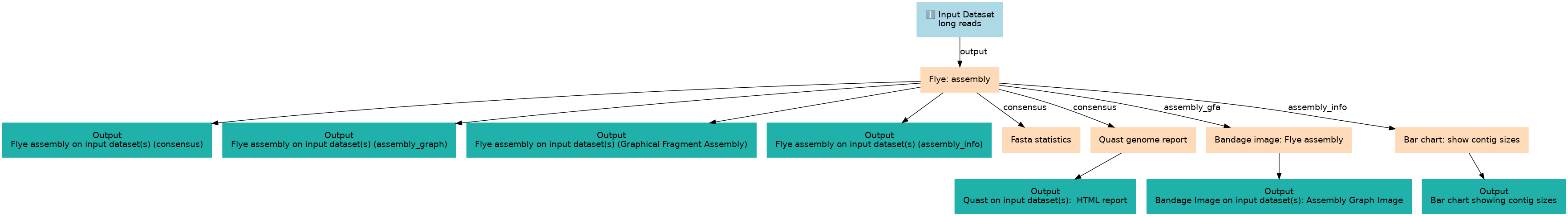
Steps
| ID | Name | Description |
|---|---|---|
| 1 | Flye: assembly | toolshed.g2.bx.psu.edu/repos/bgruening/flye/flye/2.9.3+galaxy0 |
| 2 | Fasta statistics | toolshed.g2.bx.psu.edu/repos/iuc/fasta_stats/fasta-stats/2.0 |
| 3 | Quast genome report | toolshed.g2.bx.psu.edu/repos/iuc/quast/quast/5.2.0+galaxy1 |
| 4 | Bandage image: Flye assembly | toolshed.g2.bx.psu.edu/repos/iuc/bandage/bandage_image/2022.09+galaxy4 |
| 5 | Bar chart: show contig sizes | barchart_gnuplot |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Flye assembly on input dataset(s) (consensus) | Flye assembly on input dataset(s) (consensus) | n/a |
|
| Flye assembly on input dataset(s) (assembly_graph) | Flye assembly on input dataset(s) (assembly_graph) | n/a |
|
| Flye assembly on input dataset(s) (Graphical Fragment Assembly) | Flye assembly on input dataset(s) (Graphical Fragment Assembly) | n/a |
|
| Flye assembly on input dataset(s) (assembly_info) | Flye assembly on input dataset(s) (assembly_info) | n/a |
|
| Quast on input dataset(s): HTML report | Quast on input dataset(s): HTML report | n/a |
|
| Bandage Image on input dataset(s): Assembly Graph Image | Bandage Image on input dataset(s): Assembly Graph Image | n/a |
|
| Bar chart showing contig sizes | Bar chart showing contig sizes | n/a |
|
Version History
2.0 (latest) Created 16th Jul 2024 at 14:08 by Helena Rasche
Added/updated 4 files
Open
 master
mastercac6669
4.0 (earliest) Created 25th Jun 2024 at 11:05 by Helena Rasche
Added/updated 4 files
Frozen
 4.0
4.0148c7f1
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 589 Downloads: 190
Created: 25th Jun 2024 at 11:05
Last updated: 25th Jun 2024 at 11:05
 Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992