Workflow Type: Galaxy
Open
Frozen
tutorial
Associated Tutorial
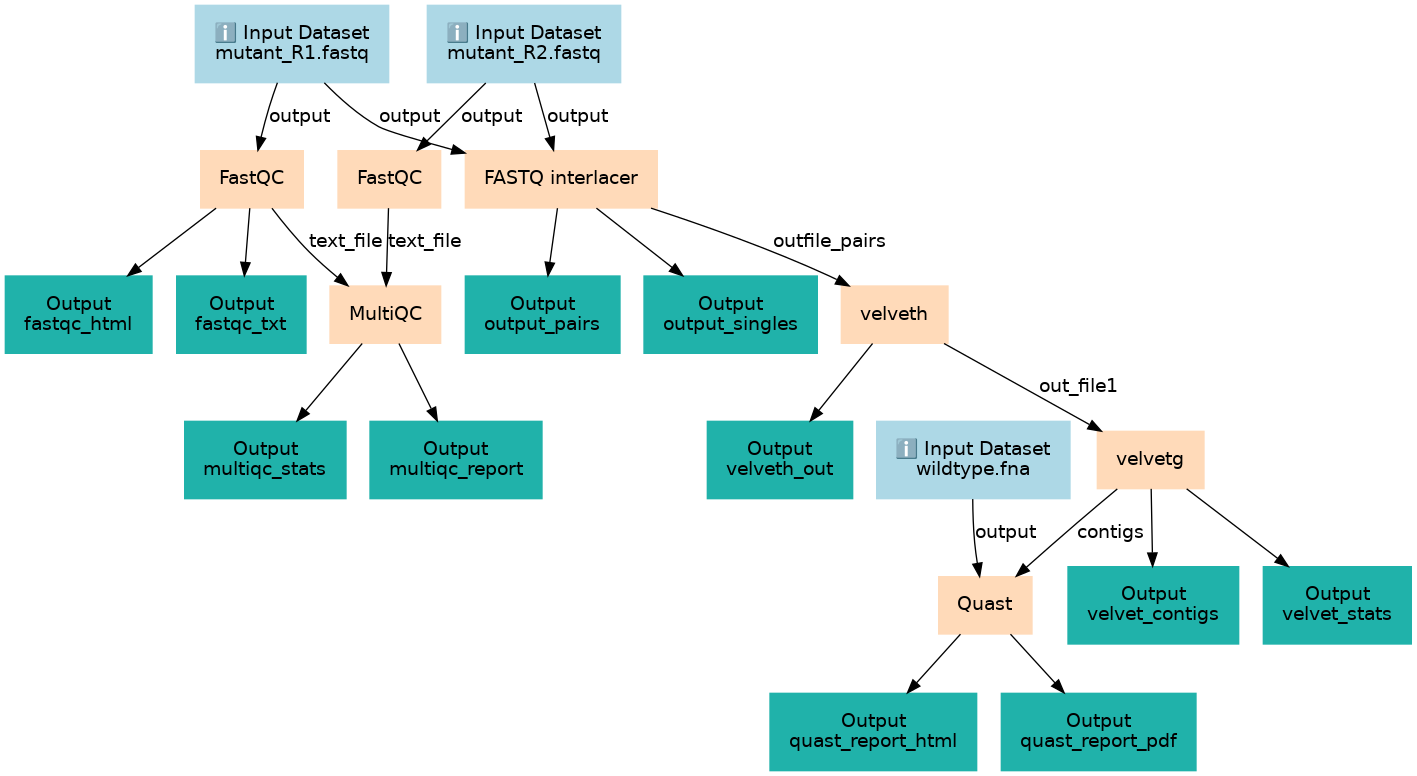
This workflows is part of the tutorial Intro to Genome Assembly, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Tutorial Author(s): Simon Gladman
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| mutant_R1.fastq | #main/mutant_R1.fastq | n/a |
|
| mutant_R2.fastq | #main/mutant_R2.fastq | n/a |
|
| wildtype.fna | #main/wildtype.fna | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 4 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 5 | FASTQ interlacer | toolshed.g2.bx.psu.edu/repos/devteam/fastq_paired_end_interlacer/fastq_paired_end_interlacer/1.2.0.1+galaxy0 |
| 6 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.7 |
| 7 | velveth | toolshed.g2.bx.psu.edu/repos/devteam/velvet/velveth/1.2.10.1 |
| 8 | velvetg | toolshed.g2.bx.psu.edu/repos/devteam/velvet/velvetg/1.2.10.1 |
| 9 | Quast | toolshed.g2.bx.psu.edu/repos/iuc/quast/quast/5.0.2+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| fastqc_html | #main/fastqc_html | n/a |
|
| fastqc_txt | #main/fastqc_txt | n/a |
|
| multiqc_report | #main/multiqc_report | n/a |
|
| multiqc_stats | #main/multiqc_stats | n/a |
|
| output_pairs | #main/output_pairs | n/a |
|
| output_singles | #main/output_singles | n/a |
|
| quast_report_html | #main/quast_report_html | n/a |
|
| quast_report_pdf | #main/quast_report_pdf | n/a |
|
| velvet_contigs | #main/velvet_contigs | n/a |
|
| velvet_stats | #main/velvet_stats | n/a |
|
| velveth_out | #main/velveth_out | n/a |
|
Version History
5.0 (latest) Created 16th Jul 2024 at 14:09 by Helena Rasche
Added/updated 4 files
Open
 master
master62dd91f
6.0 (earliest) Created 25th Jun 2024 at 11:08 by Helena Rasche
Added/updated 4 files
Frozen
 6.0
6.0b071117
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 702 Downloads: 204
Created: 25th Jun 2024 at 11:08
Last updated: 25th Jun 2024 at 11:08
 Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992