Workflow Type: Common Workflow Language
Open
Open
Open
Stable
COnSensus Interaction Network InFErence Service
Inference framework for reconstructing networks using a consensus approach between multiple methods and data sources.
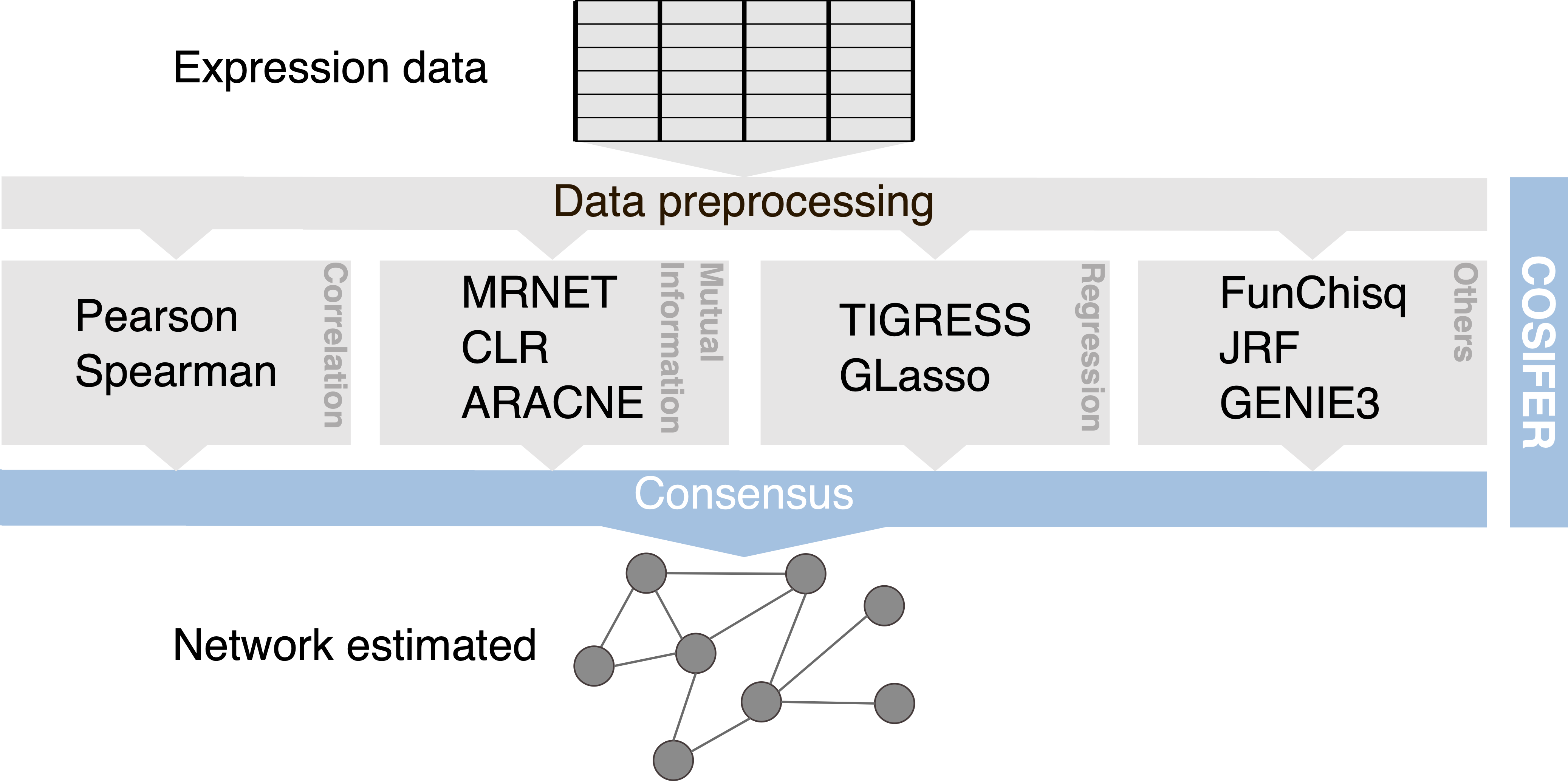
Reference
Click and drag the diagram to pan, double click or use the controls to zoom.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| data_matrix | n/a | Gene expression data matrix |
|
| gmt_filepath | n/a | Optional GMT file to perform inference on multiple gene sets |
|
| index_col | n/a | Column index in the data. Defaults to None, a.k.a., no index |
|
| outdir | n/a | Path to the output directory |
|
| separator | n/a | Separator for the data. Defaults to . |
|
| samples_on_rows | n/a | Flag that indicates that data contain the samples on rows. Defaults to False. |
|
Steps
| ID | Name | Description |
|---|---|---|
| cosifer | n/a | n/a |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| resdir | n/a | n/a |
|
Version History
Version 3 (latest) Created 16th Apr 2021 at 09:58 by Laura Rodriguez-Navas
update input descriptions
Open
 master
mastera074203
Version 2 Created 22nd Mar 2021 at 12:40 by Laura Rodriguez-Navas
No revision comments
Open
 master
masterb284735
Version 1 (earliest) Created 2nd Feb 2021 at 12:48 by Laura Rodriguez-Navas
Added/updated 1 files
Open
 master
master2970f2a
 Creators and Submitter
Creators and SubmitterCreators
Submitter
License
Activity
Views: 3278 Downloads: 686
Created: 2nd Feb 2021 at 12:48
Last updated: 16th Apr 2021 at 10:36
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub https://orcid.org/0000-0002-4806-5140
https://orcid.org/0000-0002-4806-5140