Workflow Type: Galaxy
Open
Run this on the trimmed consensus reads (DCS or SSCS) from Du Novo.
Associated Tutorial
This workflows is part of the tutorial Du Novo GTN Tutorial - Variant Calling, available in the GTN
Thanks to...
Tutorial Author(s): Anton Nekrutenko, Nick Stoler
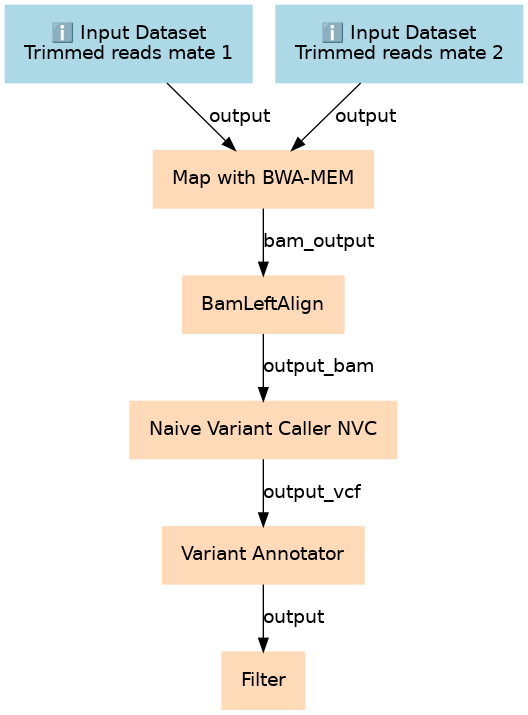
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Trimmed reads (mate 1) | #main/Trimmed reads (mate 1) | n/a |
|
| Trimmed reads (mate 2) | #main/Trimmed reads (mate 2) | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.17.1 |
| 3 | BamLeftAlign | toolshed.g2.bx.psu.edu/repos/devteam/freebayes/bamleftalign/1.1.0.46-0 |
| 4 | Naive Variant Caller (NVC) | toolshed.g2.bx.psu.edu/repos/blankenberg/naive_variant_caller/naive_variant_caller/0.0.4 |
| 5 | Variant Annotator | toolshed.g2.bx.psu.edu/repos/nick/allele_counts/allele_counts_1/1.2 |
| 6 | Filter | Filter1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
Version History
3.0 (earliest) Created 25th Jun 2024 at 10:56 by Helena Rasche
Added/updated 4 files
Open
 master
master1efd0ab
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 433 Downloads: 105
Created: 25th Jun 2024 at 10:56
Last updated: 25th Jun 2024 at 10:56
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992