Workflow Type: Galaxy
Open
Frozen
Workflows for comparison of genes in annotated genomes
Associated Tutorial
This workflows is part of the tutorial Comparative gene analysis, available in the GTN
Thanks to...
Tutorial Author(s): Anton Nekrutenko
Workflow Author(s): Anton Nekrutenko
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Diamond makedb | Diamond makedb | Diamond DB created from ORF predicted from genomes used in the analysis |
|
| Exons | Exons | Amino acid sequences of CDS exons from the gene of interest |
|
| ORFipy BED | ORFipy BED | BED dataset containing information about ORFs predicted in genomes of interest |
|
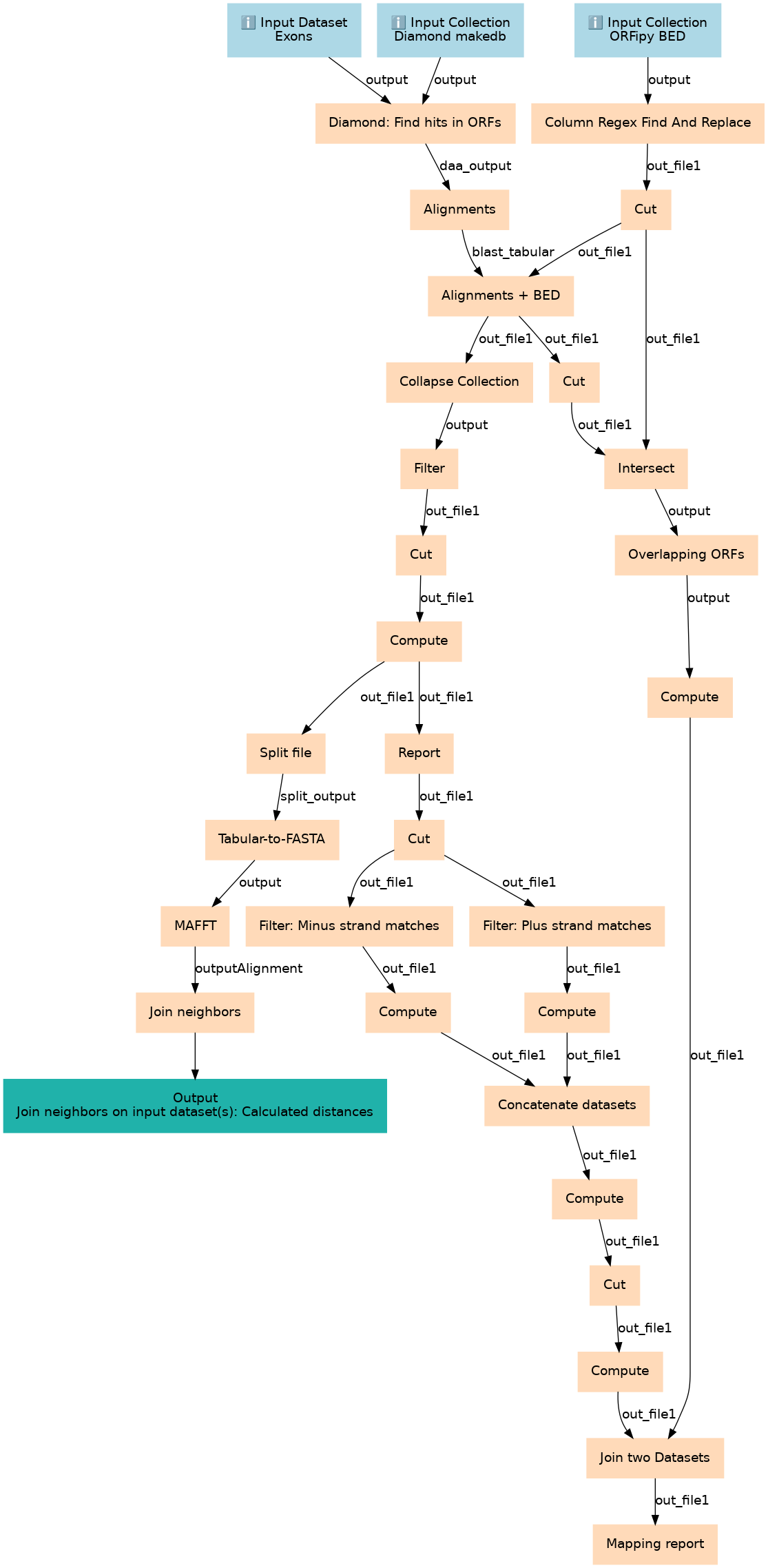
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Diamond: Find hits in ORFs | Align query against ORF translations toolshed.g2.bx.psu.edu/repos/bgruening/diamond/bg_diamond/2.0.15+galaxy0 |
| 4 | Column Regex Find And Replace | Parse name filed (column 4) pf the BED generated by ORFipy to extract name and frame information. The result has 7 columns thus is not in BED format. The next step reshuffles columns to restore BED. toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.1 |
| 5 | Alignments | Generate tabular view of alignments toolshed.g2.bx.psu.edu/repos/bgruening/diamond/bg_diamond_view/2.0.15+galaxy0 |
| 6 | Cut | Set ORF name as the name and frame as score to reestablish BED format Cut1 |
| 7 | Alignments + BED | Join tabular view of alignments with BED description of individual ORFs. This is necessary because to visualize genes we will need genomic coordinates. join1 |
| 8 | Cut | Extract genomic coordinates of matching ORFs Cut1 |
| 9 | Collapse Collection | Final list of all hits toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 10 | Intersect | Find all ORFs overlapping amino acid matches toolshed.g2.bx.psu.edu/repos/devteam/intersect/gops_intersect_1/1.0.0 |
| 11 | Filter | Filter1 |
| 12 | Overlapping ORFs | Collapse a collection into a single dataset by adding genome identified as the first column toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
| 13 | Cut | Remove unnecessary columns Cut1 |
| 14 | Compute | Create unique identified by combining genome name and the ORF name. toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 15 | Compute | Crete unique ORF id by combining genome identifier with the ORF name toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/1.6 |
| 16 | Split file | Split dataset by exon. This would create a collection in which toolshed.g2.bx.psu.edu/repos/bgruening/split_file_on_column/tp_split_on_column/0.4 |
| 17 | Report | Final textual report showing matches, their coordinates and their alignments Cut1 |
| 18 | Tabular-to-FASTA | Create amino acid FASTA sequence from aligned segments of exons toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 19 | Cut | Removing unnecessary columns for subsequent processing Cut1 |
| 20 | MAFFT | Create multiple alignments of alienable segments of axons toolshed.g2.bx.psu.edu/repos/rnateam/mafft/rbc_mafft/7.489+galaxy0 |
| 21 | Filter: Plus strand matches | Get positive strand matches Filter1 |
| 22 | Filter: Minus strand matches | Get negative strand matches Filter1 |
| 23 | Join neighbors | Compute NJ phylogenetic trees toolshed.g2.bx.psu.edu/repos/iuc/rapidnj/rapidnj/2.3.2 |
| 24 | Compute | Compute genomic coordinates of matches using global coordinates of ORFs and local coordinates of matches within ORFs toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 25 | Compute | Compute genomic coordinates of matches using global coordinates of ORFs and local coordinates of matches within ORFs toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 26 | Concatenate datasets | cat1 |
| 27 | Compute | Compute match midpoint. It is needed for creating the image. toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 28 | Cut | Cut1 |
| 29 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.0 |
| 30 | Join two Datasets | Add information about other ORFs in this area. This is done by talking all ORFs in BED format and left joining with coordinates of matched ORFs. As a result we have a sparse table that contains all ORFs surrounding our matches as well as matches themselves. This information is used to generate the final figure. join1 |
| 31 | Mapping report | Cut1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Join neighbors on input dataset(s): Calculated distances | Join neighbors on input dataset(s): Calculated distances | n/a |
|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
Version History
1.0 (latest) Created 16th Jul 2024 at 14:04 by Helena Rasche
Added/updated 4 files
Open
 master
master6c8f85e
2.0 (earliest) Created 25th Jun 2024 at 11:06 by Helena Rasche
Added/updated 4 files
Frozen
 2.0
2.04f6ea18
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 601 Downloads: 163
Created: 25th Jun 2024 at 11:06
Last updated: 25th Jun 2024 at 11:06
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992