Workflow Type: Galaxy
Open
Associated Tutorial
This workflows is part of the tutorial Cell Cycle Regression Workflow, available in the GTN
Thanks to...
Tutorial Author(s): Marisa Loach
Tutorial Contributor(s): Wendi Bacon, Graeme Tyson
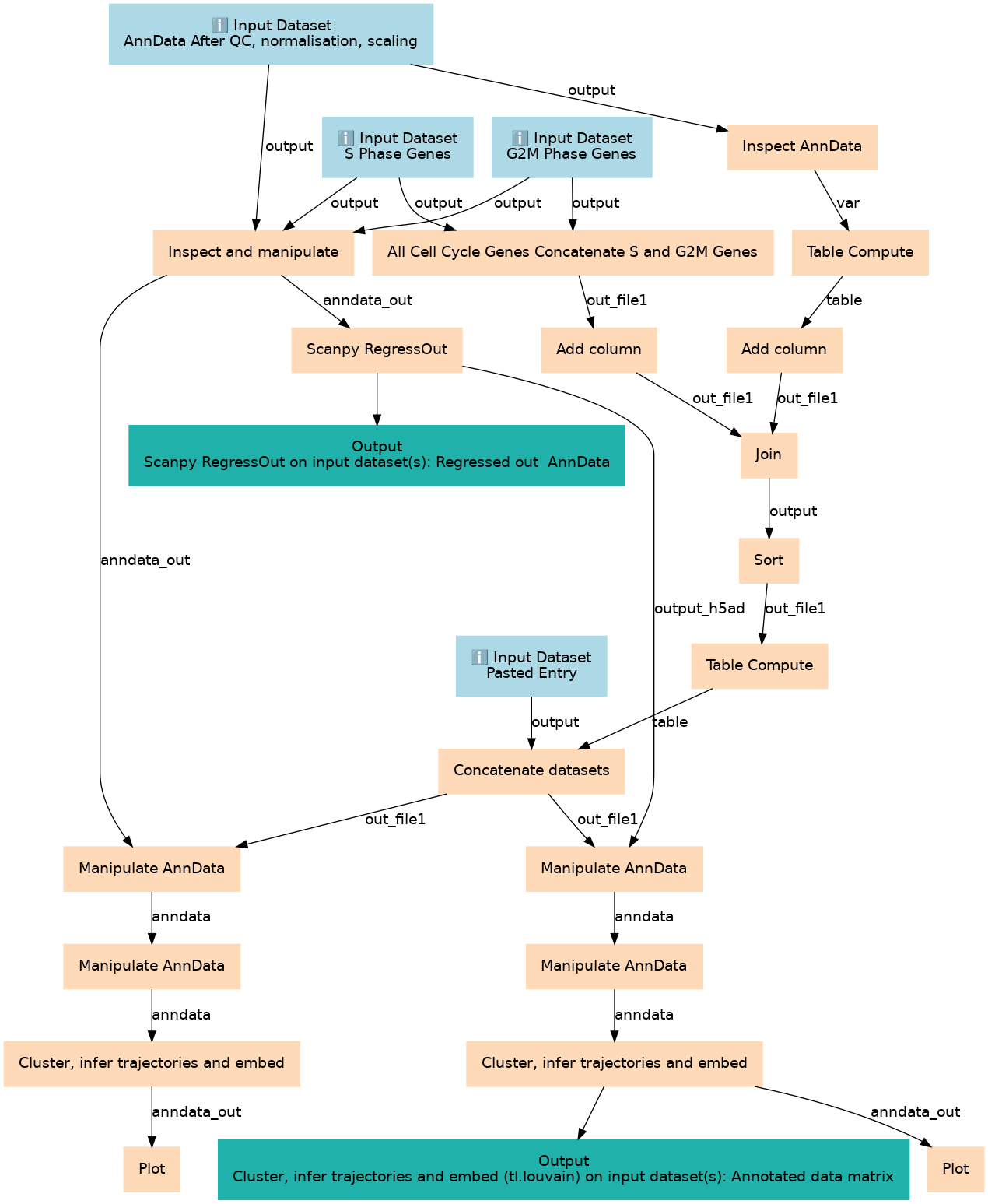
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| AnnData (After QC, normalisation, scaling) | #main/AnnData (After QC, normalisation, scaling) | n/a |
|
| G2M Phase Genes | #main/G2M Phase Genes | n/a |
|
| Pasted Entry | #main/Pasted Entry | n/a |
|
| S Phase Genes | #main/S Phase Genes | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1 |
| 5 | All Cell Cycle Genes (Concatenate S and G2M Genes) | cat1 |
| 6 | Inspect and manipulate | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.7.1+galaxy0 |
| 7 | Table Compute | toolshed.g2.bx.psu.edu/repos/iuc/table_compute/table_compute/1.2.4+galaxy0 |
| 8 | Add column | toolshed.g2.bx.psu.edu/repos/devteam/add_value/addValue/1.0.0 |
| 9 | Scanpy RegressOut | toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_regress_variable/scanpy_regress_variable/1.8.1+galaxy0 |
| 10 | Add column | toolshed.g2.bx.psu.edu/repos/devteam/add_value/addValue/1.0.0 |
| 11 | Join | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_easyjoin_tool/1.1.2 |
| 12 | Sort | sort1 |
| 13 | Table Compute | toolshed.g2.bx.psu.edu/repos/iuc/table_compute/table_compute/1.2.4+galaxy0 |
| 14 | Concatenate datasets | cat1 |
| 15 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1 |
| 16 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1 |
| 17 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1 |
| 18 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1 |
| 19 | Cluster, infer trajectories and embed | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.7.1+galaxy0 |
| 20 | Cluster, infer trajectories and embed | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.7.1+galaxy0 |
| 21 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
| 22 | Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.7.1+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Cluster, infer trajectories and embed (tl.louvain) on input dataset(s): Annotated data matrix | #main/Cluster, infer trajectories and embed (tl.louvain) on input dataset(s): Annotated data matrix | n/a |
|
| Scanpy RegressOut on input dataset(s): Regressed out AnnData | #main/Scanpy RegressOut on input dataset(s): Regressed out AnnData | n/a |
|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_11 | #main/_anonymous_output_11 | n/a |
|
| _anonymous_output_12 | #main/_anonymous_output_12 | n/a |
|
| _anonymous_output_13 | #main/_anonymous_output_13 | n/a |
|
| _anonymous_output_14 | #main/_anonymous_output_14 | n/a |
|
| _anonymous_output_15 | #main/_anonymous_output_15 | n/a |
|
| _anonymous_output_16 | #main/_anonymous_output_16 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
Version History
1.0 (earliest) Created 25th Jun 2024 at 11:15 by Helena Rasche
Added/updated 4 files
Open
 master
master2b462b8
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
Activity
Views: 413 Downloads: 100
Created: 25th Jun 2024 at 11:15
Last updated: 25th Jun 2024 at 11:15
 Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992