Workflow Type: Galaxy
Open
This workflow completes the first part of the small RNA-seq tutorial from read pre-processing to trimming.e
Associated Tutorial
This workflows is part of the tutorial sRNA Seq Step 1: Read Pre Processing And Removal Of Artifacts (no Grooming), available in the GTN
Thanks to...
Tutorial Author(s): Mallory Freeberg
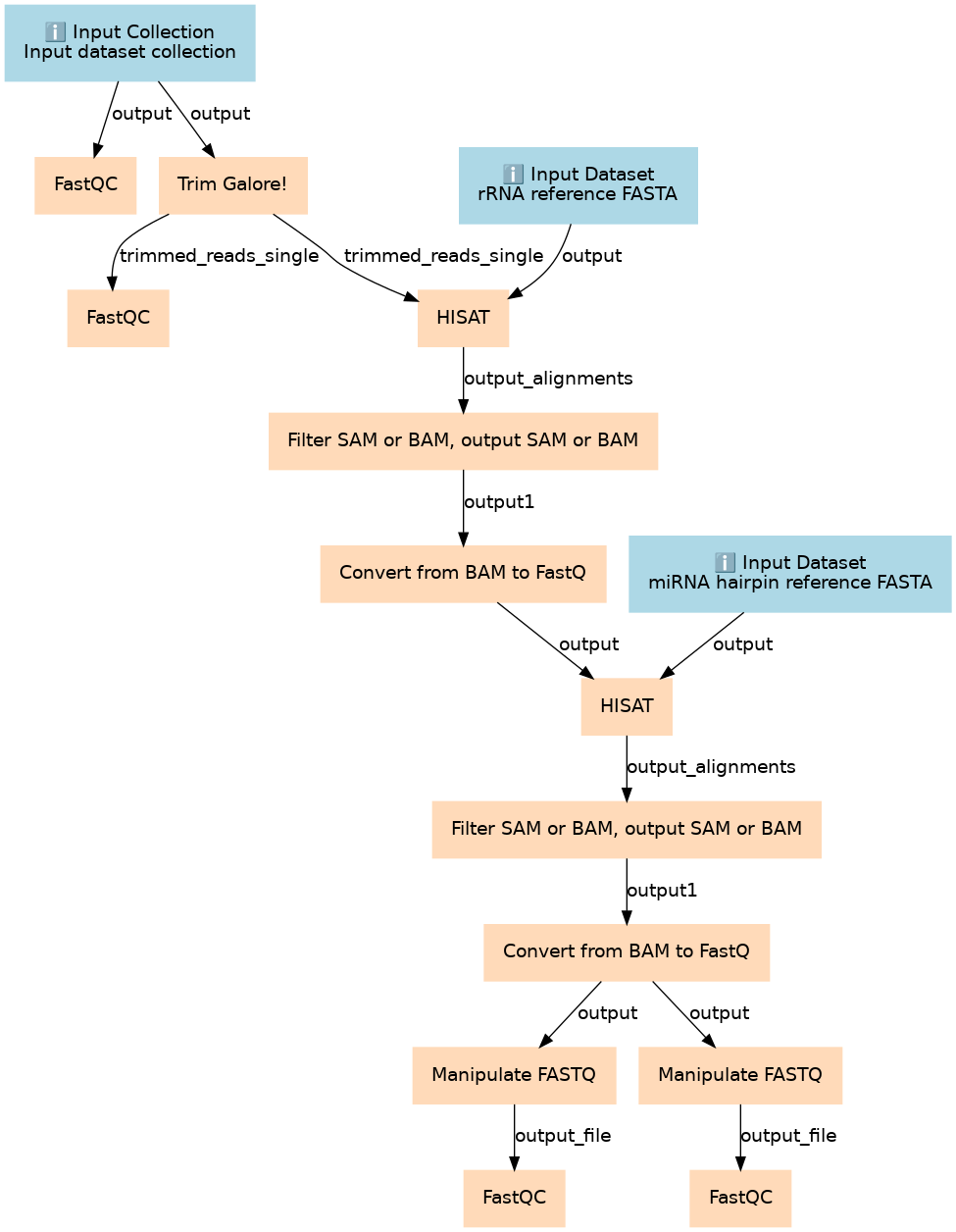
Steps
| ID | Name | Description |
|---|---|---|
| 3 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.65 |
| 4 | Trim Galore! | toolshed.g2.bx.psu.edu/repos/bgruening/trim_galore/trim_galore/0.4.2 |
| 5 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.65 |
| 6 | HISAT | Alignment of reads against rRNA sequences to filter them out. toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.0.3 |
| 7 | Filter SAM or BAM, output SAM or BAM | Keep reads that did NOT align to rRNA sequences. toolshed.g2.bx.psu.edu/repos/devteam/samtool_filter2/samtool_filter2/1.1.2 |
| 8 | Convert from BAM to FastQ | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_bamtofastq/2.24.0 |
| 9 | HISAT | toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.0.3 |
| 10 | Filter SAM or BAM, output SAM or BAM | Keep reads that did NOT align to miRNA sequences. toolshed.g2.bx.psu.edu/repos/devteam/samtool_filter2/samtool_filter2/1.1.2 |
| 11 | Convert from BAM to FastQ | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_bamtofastq/2.24.0 |
| 12 | Manipulate FASTQ | toolshed.g2.bx.psu.edu/repos/devteam/fastq_manipulation/fastq_manipulation/1.0.1 |
| 13 | Manipulate FASTQ | toolshed.g2.bx.psu.edu/repos/devteam/fastq_manipulation/fastq_manipulation/1.0.1 |
| 14 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.65 |
| 15 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.65 |
Version History
2.0 (earliest) Created 25th Jun 2024 at 11:19 by Helena Rasche
Added/updated 4 files
Open
 master
masterae9d655
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 450 Downloads: 109
Created: 25th Jun 2024 at 11:19
Last updated: 25th Jun 2024 at 11:19
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992