Workflow Type: Galaxy
Open
Frozen
Reference-based RNA-Seq data analysis
Associated Tutorial
This workflows is part of the tutorial Reference-based RNA-Seq data analysis, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Bérénice Batut, Mallory Freeberg, Mo Heydarian, Anika Erxleben, Pavankumar Videm, Clemens Blank, Maria Doyle, Nicola Soranzo, Peter van Heusden, Lucille Delisle
Tutorial Author(s): Bérénice Batut, Mallory Freeberg, Mo Heydarian, Anika Erxleben, Pavankumar Videm, Clemens Blank, Maria Doyle, Nicola Soranzo, Peter van Heusden, Lucille Delisle
Tutorial Contributor(s): Helena Rasche, Clea Siguret
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Drosophila_melanogaster.BDGP6.32.109_UCSC.gtf.gz | Drosophila_melanogaster.BDGP6.32.109_UCSC.gtf.gz | n/a |
|
| paired fastqs | paired fastqs | n/a |
|
| single fastqs | single fastqs | n/a |
|
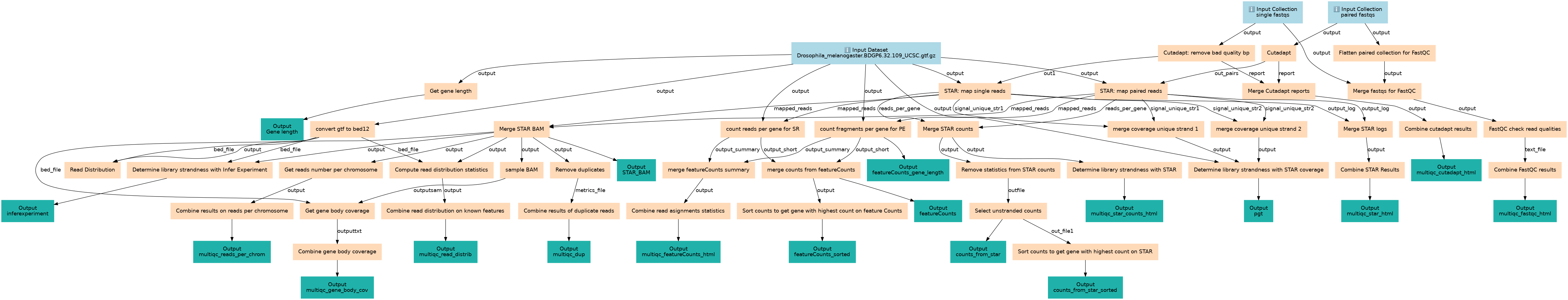
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Cutadapt: remove bad quality bp | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/4.9+galaxy1 |
| 4 | Flatten paired collection for FastQC | __FLATTEN__ |
| 5 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/4.9+galaxy1 |
| 6 | Get gene length | toolshed.g2.bx.psu.edu/repos/iuc/length_and_gc_content/length_and_gc_content/0.1.2 |
| 7 | convert gtf to bed12 | toolshed.g2.bx.psu.edu/repos/iuc/gtftobed12/gtftobed12/357 |
| 8 | STAR: map single reads | toolshed.g2.bx.psu.edu/repos/iuc/rgrnastar/rna_star/2.7.11a+galaxy0 |
| 9 | Merge fastqs for FastQC | __MERGE_COLLECTION__ |
| 10 | Merge Cutadapt reports | __MERGE_COLLECTION__ |
| 11 | STAR: map paired reads | toolshed.g2.bx.psu.edu/repos/iuc/rgrnastar/rna_star/2.7.11a+galaxy0 |
| 12 | count reads per gene for SR | toolshed.g2.bx.psu.edu/repos/iuc/featurecounts/featurecounts/2.0.3+galaxy2 |
| 13 | FastQC check read qualities | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 14 | Combine cutadapt results | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 15 | Merge STAR logs | __MERGE_COLLECTION__ |
| 16 | Merge STAR counts | __MERGE_COLLECTION__ |
| 17 | count fragments per gene for PE | toolshed.g2.bx.psu.edu/repos/iuc/featurecounts/featurecounts/2.0.3+galaxy2 |
| 18 | Merge STAR BAM | __MERGE_COLLECTION__ |
| 19 | merge coverage unique strand 1 | __MERGE_COLLECTION__ |
| 20 | merge coverage unique strand 2 | __MERGE_COLLECTION__ |
| 21 | Combine FastQC results | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 22 | Combine STAR Results | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 23 | Remove statistics from STAR counts | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_tail_tool/9.3+galaxy1 |
| 24 | Determine library strandness with STAR | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 25 | merge counts from featureCounts | __MERGE_COLLECTION__ |
| 26 | merge featureCounts summary | __MERGE_COLLECTION__ |
| 27 | Determine library strandness with Infer Experiment | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_infer_experiment/5.0.3+galaxy0 |
| 28 | Read Distribution | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_read_distribution/5.0.3+galaxy0 |
| 29 | Compute read distribution statistics | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_read_distribution/5.0.3+galaxy0 |
| 30 | sample BAM | toolshed.g2.bx.psu.edu/repos/iuc/samtools_view/samtools_view/1.15.1+galaxy2 |
| 31 | Get reads number per chromosome | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.5 |
| 32 | Remove duplicates | toolshed.g2.bx.psu.edu/repos/devteam/picard/picard_MarkDuplicates/3.1.1.0 |
| 33 | Determine library strandness with STAR coverage | toolshed.g2.bx.psu.edu/repos/iuc/pygenometracks/pygenomeTracks/3.8+galaxy2 |
| 34 | Select unstranded counts | Cut1 |
| 35 | Sort counts to get gene with highest count on feature Counts | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/9.3+galaxy1 |
| 36 | Combine read asignments statistics | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 37 | Combine read distribution on known features | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 38 | Get gene body coverage | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_geneBody_coverage/5.0.3+galaxy0 |
| 39 | Combine results on reads per chromosome | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 40 | Combine results of duplicate reads | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 41 | Sort counts to get gene with highest count on STAR | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_sort_header_tool/9.3+galaxy1 |
| 42 | Combine gene body coverage | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Gene length | Gene length | n/a |
|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| multiqc_cutadapt_html | multiqc_cutadapt_html | n/a |
|
| featureCounts_gene_length | featureCounts_gene_length | n/a |
|
| STAR_BAM | STAR_BAM | n/a |
|
| multiqc_fastqc_html | multiqc_fastqc_html | n/a |
|
| multiqc_star_html | multiqc_star_html | n/a |
|
| multiqc_star_counts_html | multiqc_star_counts_html | n/a |
|
| featureCounts | featureCounts | n/a |
|
| inferexperiment | inferexperiment | n/a |
|
| _anonymous_output_2 | _anonymous_output_2 | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| pgt | pgt | n/a |
|
| counts_from_star | counts_from_star | n/a |
|
| featureCounts_sorted | featureCounts_sorted | n/a |
|
| multiqc_featureCounts_html | multiqc_featureCounts_html | n/a |
|
| multiqc_read_distrib | multiqc_read_distrib | n/a |
|
| multiqc_reads_per_chrom | multiqc_reads_per_chrom | n/a |
|
| multiqc_dup | multiqc_dup | n/a |
|
| counts_from_star_sorted | counts_from_star_sorted | n/a |
|
| multiqc_gene_body_cov | multiqc_gene_body_cov | n/a |
|
Version History
5.0 (latest) Created 28th Oct 2024 at 13:08 by Helena Rasche
Added/updated 4 files
Open
 master
master0215b59
7.0 (earliest) Created 25th Jun 2024 at 11:25 by Helena Rasche
Added/updated 4 files
Frozen
 7.0
7.0aa7015d
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 906 Downloads: 327
Created: 25th Jun 2024 at 11:25
Last updated: 28th Oct 2024 at 13:08
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992