Workflow Type: Galaxy
Open
Frozen
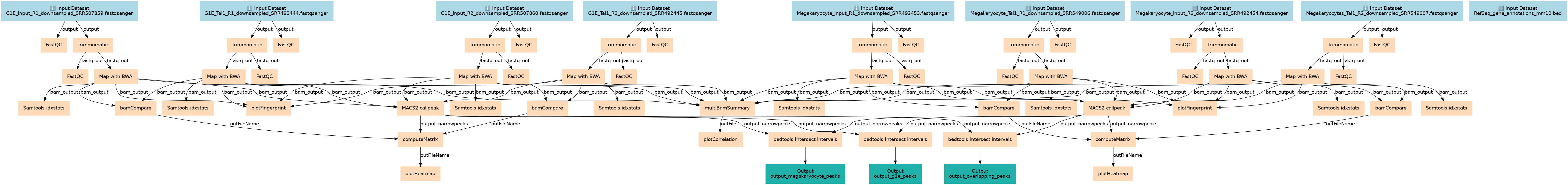
Identification of the binding sites of the T-cell acute lymphocytic leukemia protein 1 (TAL1)
Associated Tutorial
This workflows is part of the tutorial Identification of the binding sites of the T-cell acute lymphocytic leukemia protein 1 (TAL1), available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Tutorial Author(s): Mallory Freeberg, Mo Heydarian, Vivek Bhardwaj, Joachim Wolff, Anika Erxleben
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| G1E_Tal1_R1_downsampled_SRR492444.fastqsanger | G1E_Tal1_R1_downsampled_SRR492444.fastqsanger | n/a |
|
| G1E_Tal1_R2_downsampled_SRR492445.fastqsanger | G1E_Tal1_R2_downsampled_SRR492445.fastqsanger | n/a |
|
| G1E_input_R1_downsampled_SRR507859.fastqsanger | G1E_input_R1_downsampled_SRR507859.fastqsanger | n/a |
|
| G1E_input_R2_downsampled_SRR507860.fastqsanger | G1E_input_R2_downsampled_SRR507860.fastqsanger | n/a |
|
| Megakaryocyte_Tal1_R1_downsampled_SRR549006.fastqsanger | Megakaryocyte_Tal1_R1_downsampled_SRR549006.fastqsanger | n/a |
|
| Megakaryocyte_input_R1_downsampled_SRR492453.fastqsanger | Megakaryocyte_input_R1_downsampled_SRR492453.fastqsanger | n/a |
|
| Megakaryocyte_input_R2_downsampled_SRR492454.fastqsanger | Megakaryocyte_input_R2_downsampled_SRR492454.fastqsanger | n/a |
|
| Megakaryocytes_Tal1_R2_downsampled_SRR549007.fastqsanger | Megakaryocytes_Tal1_R2_downsampled_SRR549007.fastqsanger | n/a |
|
| RefSeq_gene_annotations_mm10.bed | RefSeq_gene_annotations_mm10.bed | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 9 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 10 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 11 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 12 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 13 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 14 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 15 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 16 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 17 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 18 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 19 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 20 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 21 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 22 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 23 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 24 | Trimmomatic | toolshed.g2.bx.psu.edu/repos/pjbriggs/trimmomatic/trimmomatic/0.38.0 |
| 25 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 26 | Map with BWA | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa/0.7.17.4 |
| 27 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 28 | Map with BWA | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa/0.7.17.4 |
| 29 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 30 | Map with BWA | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa/0.7.17.4 |
| 31 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 32 | Map with BWA | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa/0.7.17.4 |
| 33 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 34 | Map with BWA | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa/0.7.17.4 |
| 35 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 36 | Map with BWA | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa/0.7.17.4 |
| 37 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 38 | Map with BWA | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa/0.7.17.4 |
| 39 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 40 | Map with BWA | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa/0.7.17.4 |
| 41 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.3 |
| 42 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.3 |
| 43 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.3 |
| 44 | bamCompare | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_compare/deeptools_bam_compare/3.3.2.0.0 |
| 45 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.3 |
| 46 | plotFingerprint | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_plot_fingerprint/deeptools_plot_fingerprint/3.3.2.0.0 |
| 47 | MACS2 callpeak | toolshed.g2.bx.psu.edu/repos/iuc/macs2/macs2_callpeak/2.1.1.20160309.6 |
| 48 | bamCompare | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_compare/deeptools_bam_compare/3.3.2.0.0 |
| 49 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.3 |
| 50 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.3 |
| 51 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.3 |
| 52 | bamCompare | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_compare/deeptools_bam_compare/3.3.2.0.0 |
| 53 | Samtools idxstats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_idxstats/samtools_idxstats/2.0.3 |
| 54 | multiBamSummary | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_multi_bam_summary/deeptools_multi_bam_summary/3.3.2.0.0 |
| 55 | plotFingerprint | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_plot_fingerprint/deeptools_plot_fingerprint/3.3.2.0.0 |
| 56 | MACS2 callpeak | toolshed.g2.bx.psu.edu/repos/iuc/macs2/macs2_callpeak/2.1.1.20160309.6 |
| 57 | bamCompare | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_bam_compare/deeptools_bam_compare/3.3.2.0.0 |
| 58 | computeMatrix | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_compute_matrix/deeptools_compute_matrix/3.3.2.0.0 |
| 59 | plotCorrelation | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_plot_correlation/deeptools_plot_correlation/3.3.2.0.0 |
| 60 | bedtools Intersect intervals | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_intersectbed/2.29.0 |
| 61 | bedtools Intersect intervals | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_intersectbed/2.29.0 |
| 62 | bedtools Intersect intervals | toolshed.g2.bx.psu.edu/repos/iuc/bedtools/bedtools_intersectbed/2.29.0 |
| 63 | computeMatrix | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_compute_matrix/deeptools_compute_matrix/3.3.2.0.0 |
| 64 | plotHeatmap | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_plot_heatmap/deeptools_plot_heatmap/3.3.2.0.1 |
| 65 | plotHeatmap | toolshed.g2.bx.psu.edu/repos/bgruening/deeptools_plot_heatmap/deeptools_plot_heatmap/3.3.2.0.1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| output_overlapping_peaks | output_overlapping_peaks | n/a |
|
| output_g1e_peaks | output_g1e_peaks | n/a |
|
| output_megakaryocyte_peaks | output_megakaryocyte_peaks | n/a |
|
Version History
1.0 (latest) Created 16th Jul 2024 at 14:06 by Helena Rasche
Added/updated 4 files
Open
 master
masterf525bb2
5.0 (earliest) Created 25th Jun 2024 at 11:06 by Helena Rasche
Added/updated 4 files
Frozen
 5.0
5.047f27f4
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 607 Downloads: 189
Created: 25th Jun 2024 at 11:06
Last updated: 25th Jun 2024 at 11:06
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source
 https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992