Workflow Type: Galaxy
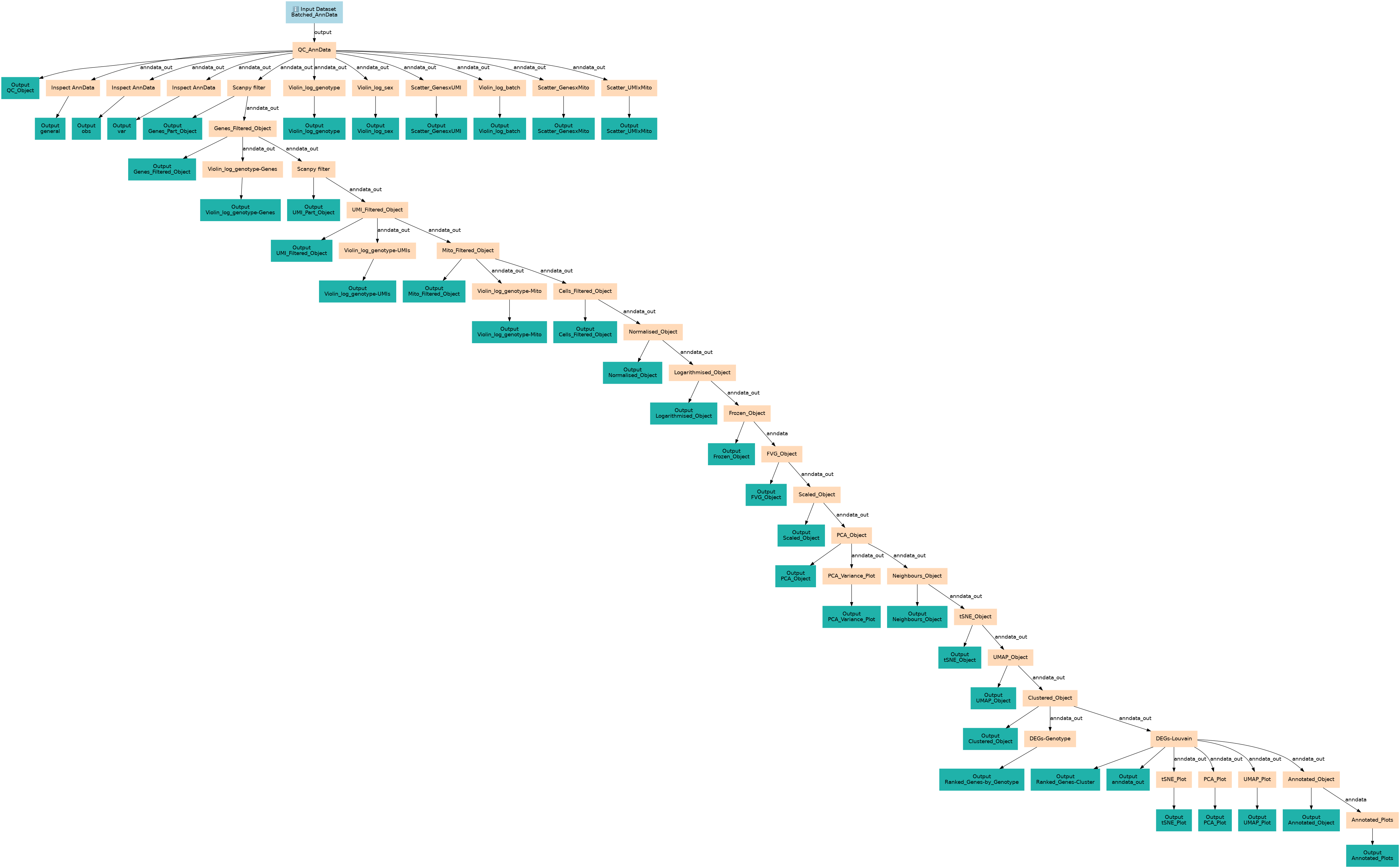
Workflow for this training: https://training.galaxyproject.org/training-material/topics/single-cell/tutorials/scrna-case_basic-pipeline/tutorial.html
Associated Tutorial
This workflows is part of the tutorial Filter, plot and explore single-cell RNA-seq data with Scanpy, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Wendi Bacon
Tutorial Author(s): Wendi Bacon
Tutorial Contributor(s): Helena Rasche, Julia Jakiela, Wendi Bacon, Pavankumar Videm, Amirhossein Naghsh Nilchi, Björn Grüning, Marisa Loach, Saskia Hiltemann, Mehmet Tekman, Pablo Moreno, Matthias Bernt, Martin Čech, David López
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Batched_AnnData | #main/Batched_AnnData | The output from this tutorial: https://training.galaxyproject.org/training-material//topics/single-cell/tutorials/scrna-case_alevin-combine-datasets/tutorial.html This is an AnnData object that has a column declaring which genes are mitochondrial or not. |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | QC_AnnData | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy2 |
| 2 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy1 |
| 3 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy1 |
| 4 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy1 |
| 5 | Scanpy filter | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy3 |
| 6 | Violin_log_genotype | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 7 | Violin_log_sex | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 8 | Scatter_GenesxUMI | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 9 | Violin_log_batch | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 10 | Scatter_GenesxMito | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 11 | Scatter_UMIxMito | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 12 | Genes_Filtered_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy3 |
| 13 | Violin_log_genotype-Genes | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 14 | Scanpy filter | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy3 |
| 15 | UMI_Filtered_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy3 |
| 16 | Violin_log_genotype-UMIs | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 17 | Mito_Filtered_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy3 |
| 18 | Violin_log_genotype-Mito | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 19 | Cells_Filtered_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy3 |
| 20 | Normalised_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_normalize/scanpy_normalize/1.10.2+galaxy0 |
| 21 | Logarithmised_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 22 | Frozen_Object | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy1 |
| 23 | FVG_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy3 |
| 24 | Scaled_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 25 | PCA_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.10.2+galaxy2 |
| 26 | PCA_Variance_Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 27 | Neighbours_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 28 | tSNE_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.10.2+galaxy2 |
| 29 | UMAP_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.10.2+galaxy2 |
| 30 | Clustered_Object | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.10.2+galaxy2 |
| 31 | DEGs-Genotype | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 32 | DEGs-Louvain | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 33 | tSNE_Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 34 | PCA_Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 35 | UMAP_Plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
| 36 | Annotated_Object | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy1 |
| 37 | Annotated_Plots | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| Annotated_Object | #main/Annotated_Object | n/a |
|
| Annotated_Plots | #main/Annotated_Plots | n/a |
|
| Cells_Filtered_Object | #main/Cells_Filtered_Object | n/a |
|
| Clustered_Object | #main/Clustered_Object | n/a |
|
| FVG_Object | #main/FVG_Object | n/a |
|
| Frozen_Object | #main/Frozen_Object | n/a |
|
| Genes_Filtered_Object | #main/Genes_Filtered_Object | n/a |
|
| Genes_Part_Object | #main/Genes_Part_Object | n/a |
|
| Logarithmised_Object | #main/Logarithmised_Object | n/a |
|
| Mito_Filtered_Object | #main/Mito_Filtered_Object | n/a |
|
| Neighbours_Object | #main/Neighbours_Object | n/a |
|
| Normalised_Object | #main/Normalised_Object | n/a |
|
| PCA_Object | #main/PCA_Object | n/a |
|
| PCA_Plot | #main/PCA_Plot | n/a |
|
| PCA_Variance_Plot | #main/PCA_Variance_Plot | n/a |
|
| QC_Object | #main/QC_Object | n/a |
|
| Ranked_Genes-Cluster | #main/Ranked_Genes-Cluster | n/a |
|
| Ranked_Genes-by_Genotype | #main/Ranked_Genes-by_Genotype | n/a |
|
| Scaled_Object | #main/Scaled_Object | n/a |
|
| Scatter_GenesxMito | #main/Scatter_GenesxMito | n/a |
|
| Scatter_GenesxUMI | #main/Scatter_GenesxUMI | n/a |
|
| Scatter_UMIxMito | #main/Scatter_UMIxMito | n/a |
|
| UMAP_Object | #main/UMAP_Object | n/a |
|
| UMAP_Plot | #main/UMAP_Plot | n/a |
|
| UMI_Filtered_Object | #main/UMI_Filtered_Object | n/a |
|
| UMI_Part_Object | #main/UMI_Part_Object | n/a |
|
| Violin_log_batch | #main/Violin_log_batch | n/a |
|
| Violin_log_genotype | #main/Violin_log_genotype | n/a |
|
| Violin_log_genotype-Genes | #main/Violin_log_genotype-Genes | n/a |
|
| Violin_log_genotype-Mito | #main/Violin_log_genotype-Mito | n/a |
|
| Violin_log_genotype-UMIs | #main/Violin_log_genotype-UMIs | n/a |
|
| Violin_log_sex | #main/Violin_log_sex | n/a |
|
| anndata_out | #main/anndata_out | n/a |
|
| general | #main/general | n/a |
|
| obs | #main/obs | n/a |
|
| tSNE_Object | #main/tSNE_Object | n/a |
|
| tSNE_Plot | #main/tSNE_Plot | n/a |
|
| var | #main/var | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 315 Downloads: 36 Runs: 0
Created: 2nd Jun 2025 at 10:56
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master