Workflow Type: Galaxy
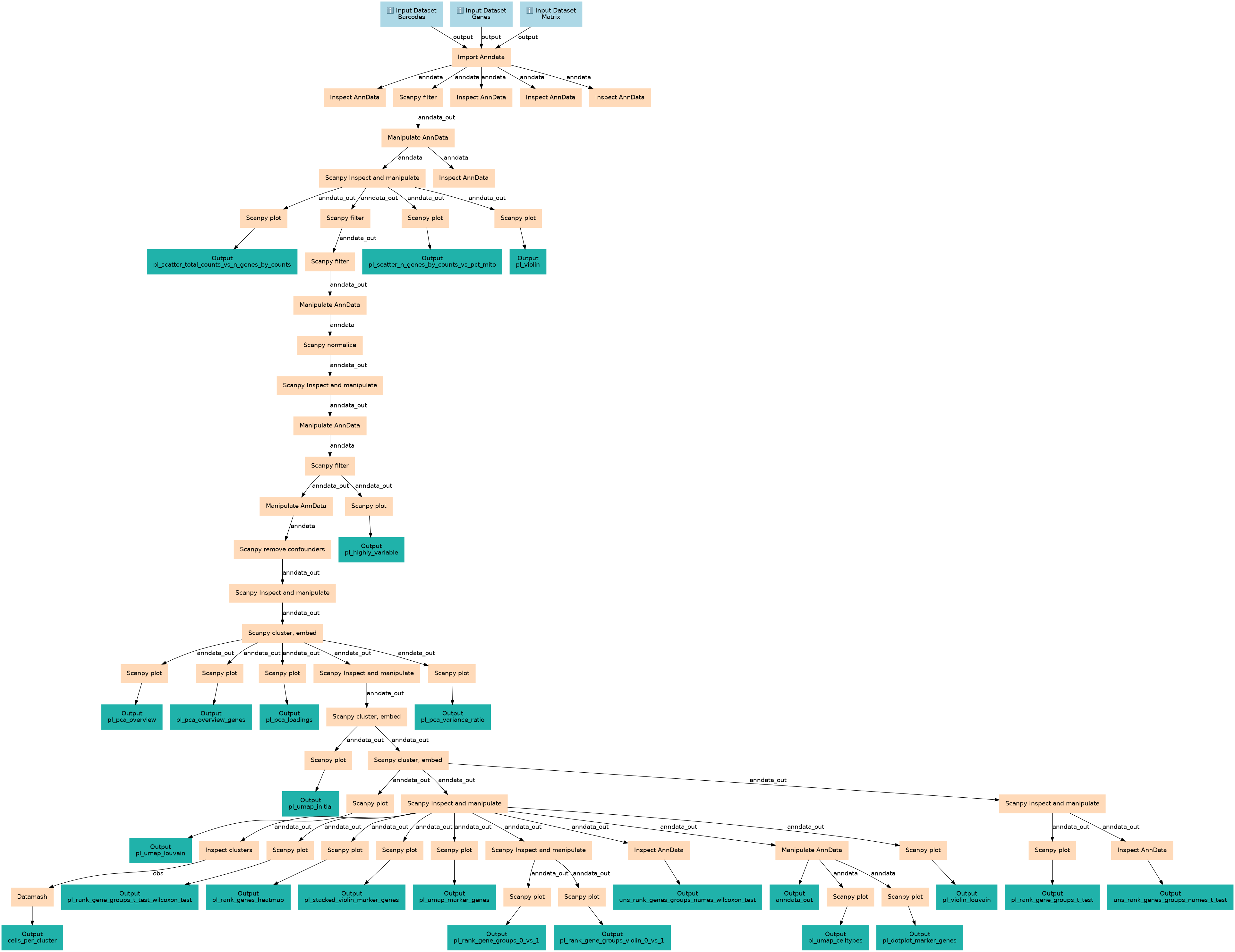
Workflow based on clustering 3K PBMCs with Scanpy tutorial
Associated Tutorial
This workflows is part of the tutorial Clustering 3K PBMCs with Scanpy, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Bérénice Batut, Hans-Rudolf Hotz, Mehmet Tekman, Pavankumar Videm
Tutorial Author(s): Bérénice Batut, Hans-Rudolf Hotz, Mehmet Tekman, Pavankumar Videm, Diana Chiang Jurado
Tutorial Contributor(s): Björn Grüning, Teresa Müller, Amirhossein Naghsh Nilchi, Saskia Hiltemann, Pavankumar Videm, Mehmet Tekman, Wendi Bacon, Helena Rasche, Hans-Rudolf Hotz, Nicola Soranzo, Marius van den Beek, Anthony Bretaudeau, Nate Coraor
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Barcodes | Barcodes | n/a |
|
| Genes | Genes | n/a |
|
| Matrix | Matrix | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Import Anndata | toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0 |
| 4 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0 |
| 5 | Scanpy filter | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy0 |
| 6 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0 |
| 7 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0 |
| 8 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0 |
| 9 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0 |
| 10 | Scanpy Inspect and manipulate | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 11 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0 |
| 12 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 13 | Scanpy filter | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy0 |
| 14 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 15 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 16 | Scanpy filter | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy0 |
| 17 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0 |
| 18 | Scanpy normalize | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_normalize/scanpy_normalize/1.10.2+galaxy0 |
| 19 | Scanpy Inspect and manipulate | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 20 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0 |
| 21 | Scanpy filter | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_filter/scanpy_filter/1.10.2+galaxy0 |
| 22 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0 |
| 23 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 24 | Scanpy remove confounders | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_remove_confounders/scanpy_remove_confounders/1.10.2+galaxy0 |
| 25 | Scanpy Inspect and manipulate | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 26 | Scanpy cluster, embed | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.10.2+galaxy0 |
| 27 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 28 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 29 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 30 | Scanpy Inspect and manipulate | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 31 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 32 | Scanpy cluster, embed | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.10.2+galaxy0 |
| 33 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 34 | Scanpy cluster, embed | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_cluster_reduce_dimension/scanpy_cluster_reduce_dimension/1.10.2+galaxy0 |
| 35 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 36 | Scanpy Inspect and manipulate | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 37 | Scanpy Inspect and manipulate | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 38 | Inspect clusters | Here I inspected the louviain clusters... toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0 |
| 39 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 40 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 41 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 42 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 43 | Scanpy Inspect and manipulate | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_inspect/scanpy_inspect/1.10.2+galaxy1 |
| 44 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0 |
| 45 | Manipulate AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0 |
| 46 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 47 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 48 | Inspect AnnData | toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0 |
| 49 | Datamash | toolshed.g2.bx.psu.edu/repos/iuc/datamash_ops/datamash_ops/1.8+galaxy0 |
| 50 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 51 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 52 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
| 53 | Scanpy plot | toolshed.g2.bx.psu.edu/repos/iuc/scanpy_plot/scanpy_plot/1.10.2+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| pl_scatter_total_counts_vs_n_genes_by_counts | pl_scatter_total_counts_vs_n_genes_by_counts | n/a |
|
| pl_scatter_n_genes_by_counts_vs_pct_mito | pl_scatter_n_genes_by_counts_vs_pct_mito | n/a |
|
| pl_violin | pl_violin | n/a |
|
| pl_highly_variable | pl_highly_variable | n/a |
|
| pl_pca_overview | pl_pca_overview | n/a |
|
| pl_pca_overview_genes | pl_pca_overview_genes | n/a |
|
| pl_pca_loadings | pl_pca_loadings | n/a |
|
| pl_pca_variance_ratio | pl_pca_variance_ratio | n/a |
|
| pl_umap_initial | pl_umap_initial | n/a |
|
| pl_umap_louvain | pl_umap_louvain | n/a |
|
| pl_rank_gene_groups_t_test_wilcoxon_test | pl_rank_gene_groups_t_test_wilcoxon_test | n/a |
|
| pl_rank_genes_heatmap | pl_rank_genes_heatmap | n/a |
|
| pl_stacked_violin_marker_genes | pl_stacked_violin_marker_genes | n/a |
|
| pl_umap_marker_genes | pl_umap_marker_genes | n/a |
|
| uns_rank_genes_groups_names_wilcoxon_test | uns_rank_genes_groups_names_wilcoxon_test | n/a |
|
| anndata_out | anndata_out | n/a |
|
| pl_violin_louvain | pl_violin_louvain | n/a |
|
| pl_rank_gene_groups_t_test | pl_rank_gene_groups_t_test | n/a |
|
| uns_rank_genes_groups_names_t_test | uns_rank_genes_groups_names_t_test | n/a |
|
| cells_per_cluster | cells_per_cluster | n/a |
|
| pl_rank_gene_groups_0_vs_1 | pl_rank_gene_groups_0_vs_1 | n/a |
|
| pl_rank_gene_groups_violin_0_vs_1 | pl_rank_gene_groups_violin_0_vs_1 | n/a |
|
| pl_umap_celltypes | pl_umap_celltypes | n/a |
|
| pl_dotplot_marker_genes | pl_dotplot_marker_genes | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 247 Downloads: 56 Runs: 0
Created: 2nd Jun 2025 at 10:56
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master