Workflow Type: Galaxy
Associated Tutorial
This workflows is part of the tutorial Using BioImage.IO models for image analysis in Galaxy, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Diana Chiang Jurado, Leonid Kostrykin
Tutorial Author(s): Diana Chiang Jurado, Leonid Kostrykin
Tutorial Contributor(s): Leonid Kostrykin, Saskia Hiltemann, Diana Chiang Jurado, Björn Grüning, Beatriz Serrano-Solano, Anup Kumar
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| BioImage.IO Model | BioImage.IO Model | This PyTorch [.pt] file is located in our Galaxy repository under the ML model/bioimaging-models directory. |
|
| Image for Prediction | Image for Prediction | Input image required for inference with the ML model. Ensure that the image is of the same biological type the model was trained on or is designed to process. The tool only accepts image files in TIFF or PNG format. |
|
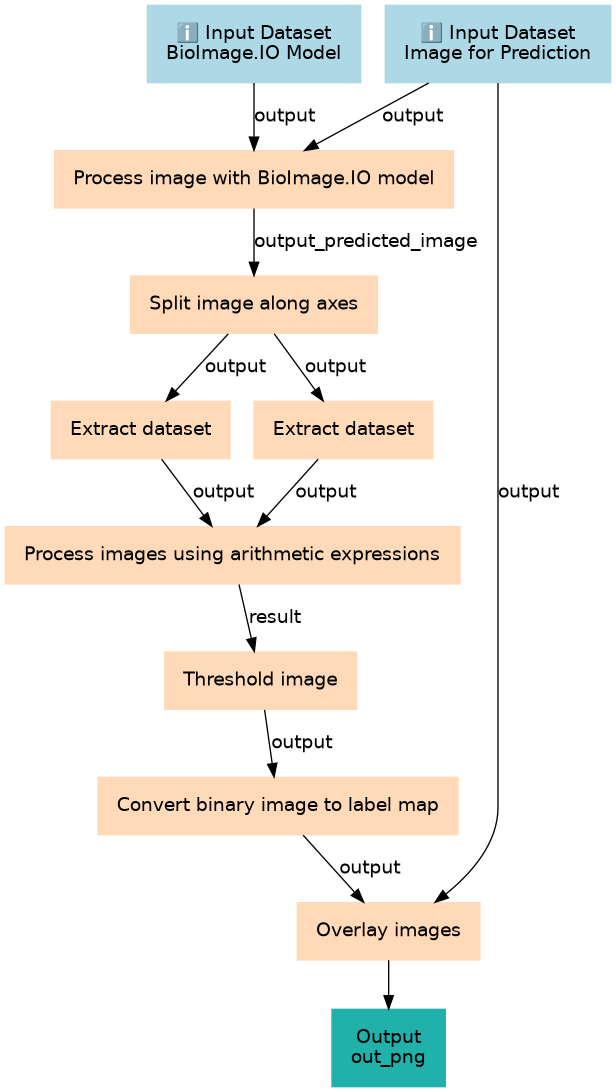
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Process image with BioImage.IO model | toolshed.g2.bx.psu.edu/repos/bgruening/bioimage_inference/bioimage_inference/2.4.1+galaxy3 |
| 3 | Split image along axes | toolshed.g2.bx.psu.edu/repos/imgteam/split_image/ip_split_image/2.2.3+galaxy1 |
| 4 | Extract dataset | __EXTRACT_DATASET__ |
| 5 | Extract dataset | __EXTRACT_DATASET__ |
| 6 | Process images using arithmetic expressions | toolshed.g2.bx.psu.edu/repos/imgteam/image_math/image_math/1.26.4+galaxy2 |
| 7 | Threshold image | toolshed.g2.bx.psu.edu/repos/imgteam/2d_auto_threshold/ip_threshold/0.18.1+galaxy3 |
| 8 | Convert binary image to label map | toolshed.g2.bx.psu.edu/repos/imgteam/binary2labelimage/ip_binary_to_labelimage/0.5+galaxy0 |
| 9 | Overlay images | toolshed.g2.bx.psu.edu/repos/imgteam/overlay_images/ip_overlay_images/0.0.4+galaxy4 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| out_png | out_png | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 286 Downloads: 34 Runs: 0
Created: 2nd Jun 2025 at 10:57
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master