Workflow Type: Galaxy
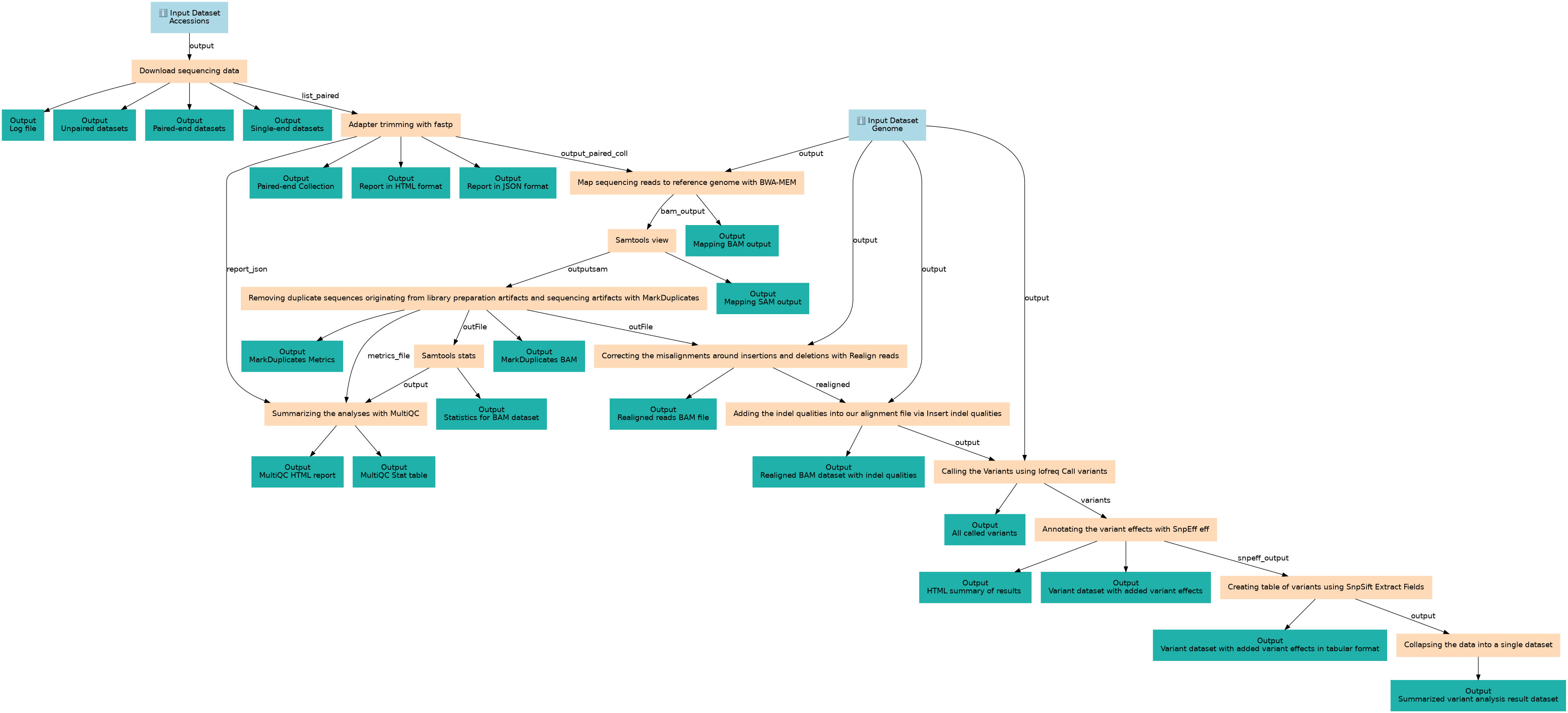
NGS data logistics
Associated Tutorial
This workflows is part of the tutorial NGS data logistics, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): Anton Nekrutenko, Marius van den Beek, Dave Clements, Daniel Blankenberg, Armin Dadras
Tutorial Author(s): Anton Nekrutenko, Marius van den Beek, Dave Clements, Daniel Blankenberg
Tutorial Contributor(s): Armin Dadras, Helena Rasche, Saskia Hiltemann, Mehmet Tekman, Nicola Soranzo, Björn Grüning, Anton Nekrutenko, John Davis, William Durand, Bérénice Batut, Niall Beard, Teresa Müller
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Accessions | #main/Accessions | Short Read Archive (SRA) accession to be downloaded using the fasterq-dump utility of the SRA Toolkit from the National Center for Biotechnology Information (NCBI). |
|
| Genome | #main/Genome | The genome that reads will be mapped to it. |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Download sequencing data | toolshed.g2.bx.psu.edu/repos/iuc/sra_tools/fasterq_dump/3.1.1+galaxy1 |
| 3 | Adapter trimming with fastp | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.24.0+galaxy4 |
| 4 | Map sequencing reads to reference genome with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.19 |
| 5 | Samtools view | toolshed.g2.bx.psu.edu/repos/iuc/samtools_view/samtools_view/1.20+galaxy3 |
| 6 | Removing duplicate sequences originating from library preparation artifacts and sequencing artifacts with MarkDuplicates | toolshed.g2.bx.psu.edu/repos/devteam/picard/picard_MarkDuplicates/3.1.1.0 |
| 7 | Correcting the misalignments around insertions and deletions with Realign reads | toolshed.g2.bx.psu.edu/repos/iuc/lofreq_viterbi/lofreq_viterbi/2.1.5+galaxy0 |
| 8 | Samtools stats | toolshed.g2.bx.psu.edu/repos/devteam/samtools_stats/samtools_stats/2.0.5 |
| 9 | Adding the indel qualities into our alignment file via Insert indel qualities | toolshed.g2.bx.psu.edu/repos/iuc/lofreq_indelqual/lofreq_indelqual/2.1.5+galaxy1 |
| 10 | Summarizing the analyses with MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.27+galaxy3 |
| 11 | Calling the Variants using lofreq Call variants | toolshed.g2.bx.psu.edu/repos/iuc/lofreq_call/lofreq_call/2.1.5+galaxy3 |
| 12 | Annotating the variant effects with SnpEff eff | toolshed.g2.bx.psu.edu/repos/iuc/snpeff_sars_cov_2/snpeff_sars_cov_2/4.5covid19 |
| 13 | Creating table of variants using SnpSift Extract Fields | toolshed.g2.bx.psu.edu/repos/iuc/snpsift/snpSift_extractFields/4.3+t.galaxy0 |
| 14 | Collapsing the data into a single dataset | toolshed.g2.bx.psu.edu/repos/nml/collapse_collections/collapse_dataset/5.1.0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| All called variants | #main/All called variants | n/a |
|
| HTML summary of results | #main/HTML summary of results | n/a |
|
| Log file | #main/Log file | n/a |
|
| Mapping BAM output | #main/Mapping BAM output | n/a |
|
| Mapping SAM output | #main/Mapping SAM output | n/a |
|
| MarkDuplicates BAM | #main/MarkDuplicates BAM | n/a |
|
| MarkDuplicates Metrics | #main/MarkDuplicates Metrics | n/a |
|
| MultiQC HTML report | #main/MultiQC HTML report | n/a |
|
| MultiQC Stat table | #main/MultiQC Stat table | n/a |
|
| Paired-end Collection | #main/Paired-end Collection | n/a |
|
| Paired-end datasets | #main/Paired-end datasets | n/a |
|
| Realigned BAM dataset with indel qualities | #main/Realigned BAM dataset with indel qualities | n/a |
|
| Realigned reads BAM file | #main/Realigned reads BAM file | n/a |
|
| Report in HTML format | #main/Report in HTML format | n/a |
|
| Report in JSON format | #main/Report in JSON format | n/a |
|
| Single-end datasets | #main/Single-end datasets | n/a |
|
| Statistics for BAM dataset | #main/Statistics for BAM dataset | n/a |
|
| Summarized variant analysis result dataset | #main/Summarized variant analysis result dataset | n/a |
|
| Unpaired datasets | #main/Unpaired datasets | n/a |
|
| Variant dataset with added variant effects | #main/Variant dataset with added variant effects | n/a |
|
| Variant dataset with added variant effects in tabular format | #main/Variant dataset with added variant effects in tabular format | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 302 Downloads: 33 Runs: 0
Created: 2nd Jun 2025 at 10:57
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master