Workflow Type: Galaxy
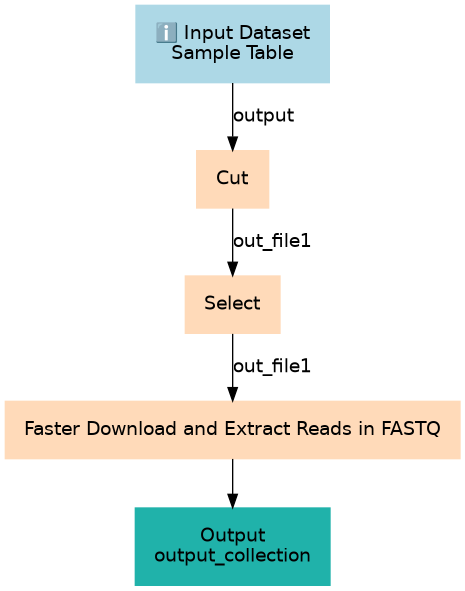
Downloads data from NCBI assuming the correct data input format. (I.e. it should be two columns, with a header, Run and Group, where Run is an SRR identifier.)
Associated Tutorial
This workflows is part of the tutorial Pathway analysis with the MINERVA Platform, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Helena Rasche
Tutorial Author(s): Marek Ostaszewski, Matti Hoch, Iacopo Cristoferi, Myrthe van Baardwijk, Saskia Hiltemann, Helena Rasche
Tutorial Contributor(s): Helena Rasche, Matti Hoch, Mira Kuntz, José Manuel Domínguez, Sanjay Kumar Srikakulam, Björn Grüning, Linelle Abueg, Saskia Hiltemann
Grants(s): BeYond-COVID
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Sample Table | #main/Sample Table | This should be a two column sample table that looks like: ``` Run Group SRR16681566 healthy SRR16681565 healthy SRR16681564 healthy ``` |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | Cut | Cut1 |
| 2 | Select | Grep1 |
| 3 | Faster Download and Extract Reads in FASTQ | toolshed.g2.bx.psu.edu/repos/iuc/sra_tools/fasterq_dump/3.0.8+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| output_collection | #main/output_collection | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
Activity
Views: 305 Downloads: 35 Runs: 0
Created: 2nd Jun 2025 at 11:04
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master