Workflow Type: Galaxy
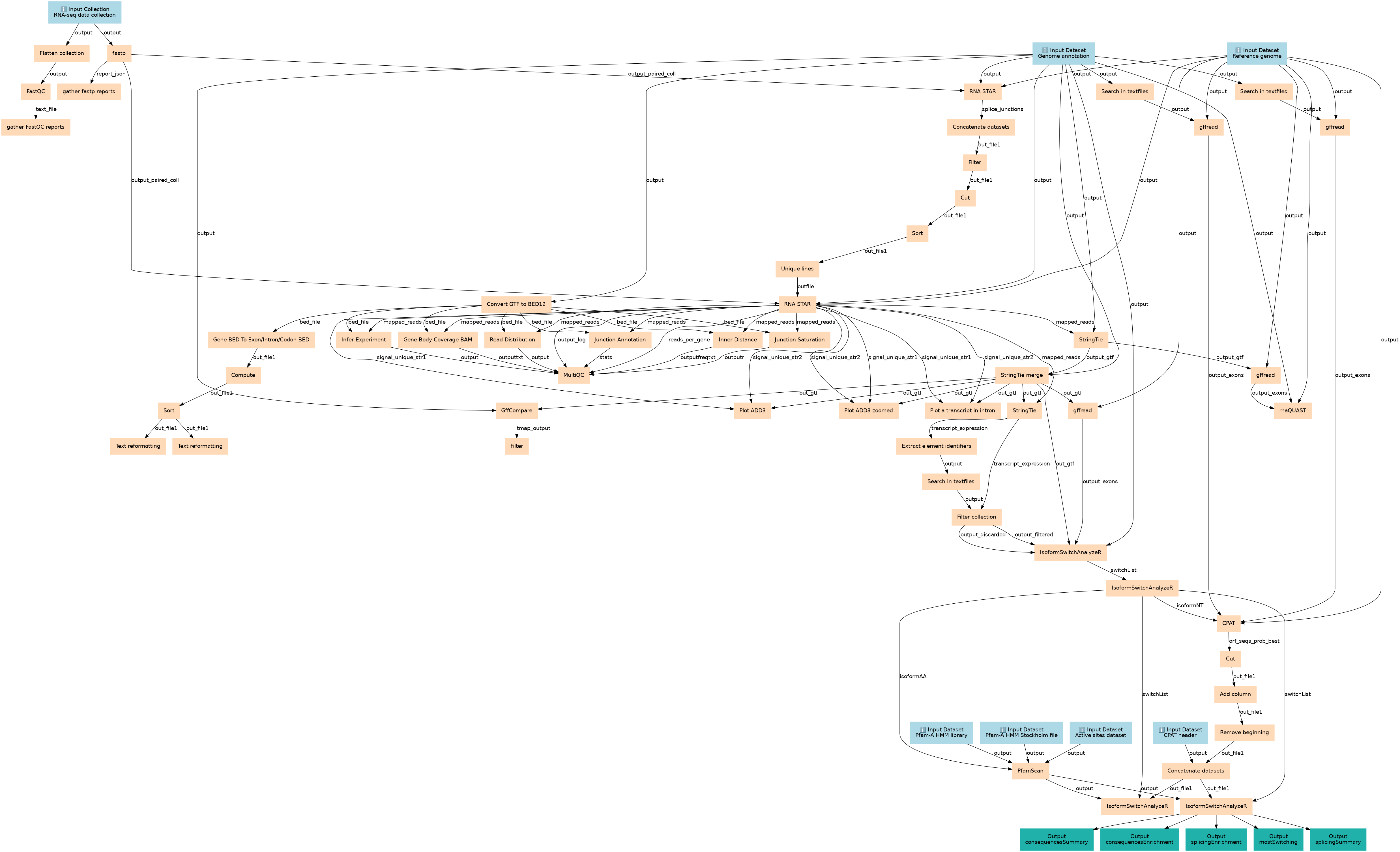
Analyze differential isoform expression
Associated Tutorial
This workflows is part of the tutorial Genome-wide alternative splicing analysis, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Cristobal Gallardo, Lucille Delisle
Tutorial Author(s): Cristóbal Gallardo, Lucille Delisle
Tutorial Contributor(s): Pavankumar Videm, Saskia Hiltemann, Björn Grüning, Lucille Delisle, Cristóbal Gallardo, Helena Rasche, Wolfgang Maier
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Active sites dataset | Active sites dataset | active_site.dat.gz from EMBL-EBI |
|
| CPAT header | CPAT header | header |
|
| Genome annotation | Genome annotation | gene annotation file |
|
| Pfam-A HMM Stockholm file | Pfam-A HMM Stockholm file | Pfam-A.hmm.dat.gz from EMBL-EBI |
|
| Pfam-A HMM library | Pfam-A HMM library | Pfam-A.hmm.gz from EMBL-EBI |
|
| RNA-seq data collection | RNA-seq data collection | input fastqs |
|
| Reference genome | Reference genome | genome reference fasta file |
|
Steps
| ID | Name | Description |
|---|---|---|
| 7 | Flatten collection | __FLATTEN__ |
| 8 | fastp | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.23.4+galaxy1 |
| 9 | Convert GTF to BED12 | toolshed.g2.bx.psu.edu/repos/iuc/gtftobed12/gtftobed12/357 |
| 10 | Search in textfiles | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy1 |
| 11 | Search in textfiles | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy1 |
| 12 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy1 |
| 13 | gather fastp reports | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 14 | RNA STAR | toolshed.g2.bx.psu.edu/repos/iuc/rgrnastar/rna_star/2.7.11a+galaxy1 |
| 15 | Gene BED To Exon/Intron/Codon BED | gene2exon1 |
| 16 | gffread | toolshed.g2.bx.psu.edu/repos/devteam/gffread/gffread/2.2.1.4+galaxy0 |
| 17 | gffread | toolshed.g2.bx.psu.edu/repos/devteam/gffread/gffread/2.2.1.4+galaxy0 |
| 18 | gather FastQC reports | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 19 | Concatenate datasets | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cat/9.3+galaxy1 |
| 20 | Compute | toolshed.g2.bx.psu.edu/repos/devteam/column_maker/Add_a_column1/2.1 |
| 21 | Filter | Filter1 |
| 22 | Sort | sort1 |
| 23 | Cut | Cut1 |
| 24 | Text reformatting | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy1 |
| 25 | Text reformatting | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/9.3+galaxy1 |
| 26 | Sort | sort1 |
| 27 | Unique lines | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_uniq_tool/9.3+galaxy1 |
| 28 | RNA STAR | toolshed.g2.bx.psu.edu/repos/iuc/rgrnastar/rna_star/2.7.11a+galaxy1 |
| 29 | StringTie | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie/2.2.3+galaxy0 |
| 30 | Infer Experiment | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_infer_experiment/5.0.3+galaxy0 |
| 31 | Gene Body Coverage (BAM) | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_geneBody_coverage/5.0.3+galaxy0 |
| 32 | Junction Saturation | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_junction_saturation/5.0.3+galaxy0 |
| 33 | Junction Annotation | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_junction_annotation/5.0.3+galaxy0 |
| 34 | Inner Distance | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_inner_distance/5.0.3+galaxy0 |
| 35 | Read Distribution | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_read_distribution/5.0.3+galaxy0 |
| 36 | StringTie merge | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie_merge/2.2.3+galaxy0 |
| 37 | gffread | toolshed.g2.bx.psu.edu/repos/devteam/gffread/gffread/2.2.1.4+galaxy0 |
| 38 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
| 39 | StringTie | toolshed.g2.bx.psu.edu/repos/iuc/stringtie/stringtie/2.2.3+galaxy0 |
| 40 | GffCompare | toolshed.g2.bx.psu.edu/repos/iuc/gffcompare/gffcompare/0.12.6+galaxy0 |
| 41 | Plot a transcript in intron | toolshed.g2.bx.psu.edu/repos/iuc/pygenometracks/pygenomeTracks/3.8+galaxy2 |
| 42 | gffread | toolshed.g2.bx.psu.edu/repos/devteam/gffread/gffread/2.2.1.4+galaxy0 |
| 43 | Plot ADD3 | toolshed.g2.bx.psu.edu/repos/iuc/pygenometracks/pygenomeTracks/3.8+galaxy2 |
| 44 | Plot ADD3 zoomed | toolshed.g2.bx.psu.edu/repos/iuc/pygenometracks/pygenomeTracks/3.8+galaxy2 |
| 45 | rnaQUAST | toolshed.g2.bx.psu.edu/repos/iuc/rnaquast/rna_quast/2.2.3+galaxy0 |
| 46 | Extract element identifiers | toolshed.g2.bx.psu.edu/repos/iuc/collection_element_identifiers/collection_element_identifiers/0.0.2 |
| 47 | Filter | Filter1 |
| 48 | Search in textfiles | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_grep_tool/9.3+galaxy1 |
| 49 | Filter collection | __FILTER_FROM_FILE__ |
| 50 | IsoformSwitchAnalyzeR | toolshed.g2.bx.psu.edu/repos/iuc/isoformswitchanalyzer/isoformswitchanalyzer/1.20.0+galaxy5 |
| 51 | IsoformSwitchAnalyzeR | toolshed.g2.bx.psu.edu/repos/iuc/isoformswitchanalyzer/isoformswitchanalyzer/1.20.0+galaxy5 |
| 52 | PfamScan | toolshed.g2.bx.psu.edu/repos/bgruening/pfamscan/pfamscan/1.6+galaxy0 |
| 53 | CPAT | toolshed.g2.bx.psu.edu/repos/bgruening/cpat/cpat/3.0.5+galaxy1 |
| 54 | Cut | Cut1 |
| 55 | Add column | toolshed.g2.bx.psu.edu/repos/devteam/add_value/addValue/1.0.1 |
| 56 | Remove beginning | Remove beginning1 |
| 57 | Concatenate datasets | cat1 |
| 58 | IsoformSwitchAnalyzeR | toolshed.g2.bx.psu.edu/repos/iuc/isoformswitchanalyzer/isoformswitchanalyzer/1.20.0+galaxy5 |
| 59 | IsoformSwitchAnalyzeR | toolshed.g2.bx.psu.edu/repos/iuc/isoformswitchanalyzer/isoformswitchanalyzer/1.20.0+galaxy5 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| consequencesSummary | consequencesSummary | n/a |
|
| consequencesEnrichment | consequencesEnrichment | n/a |
|
| splicingEnrichment | splicingEnrichment | n/a |
|
| mostSwitching | mostSwitching | n/a |
|
| splicingSummary | splicingSummary | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
License
Activity
Views: 314 Downloads: 41 Runs: 0
Created: 2nd Jun 2025 at 11:04
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master