Workflow Type: Galaxy
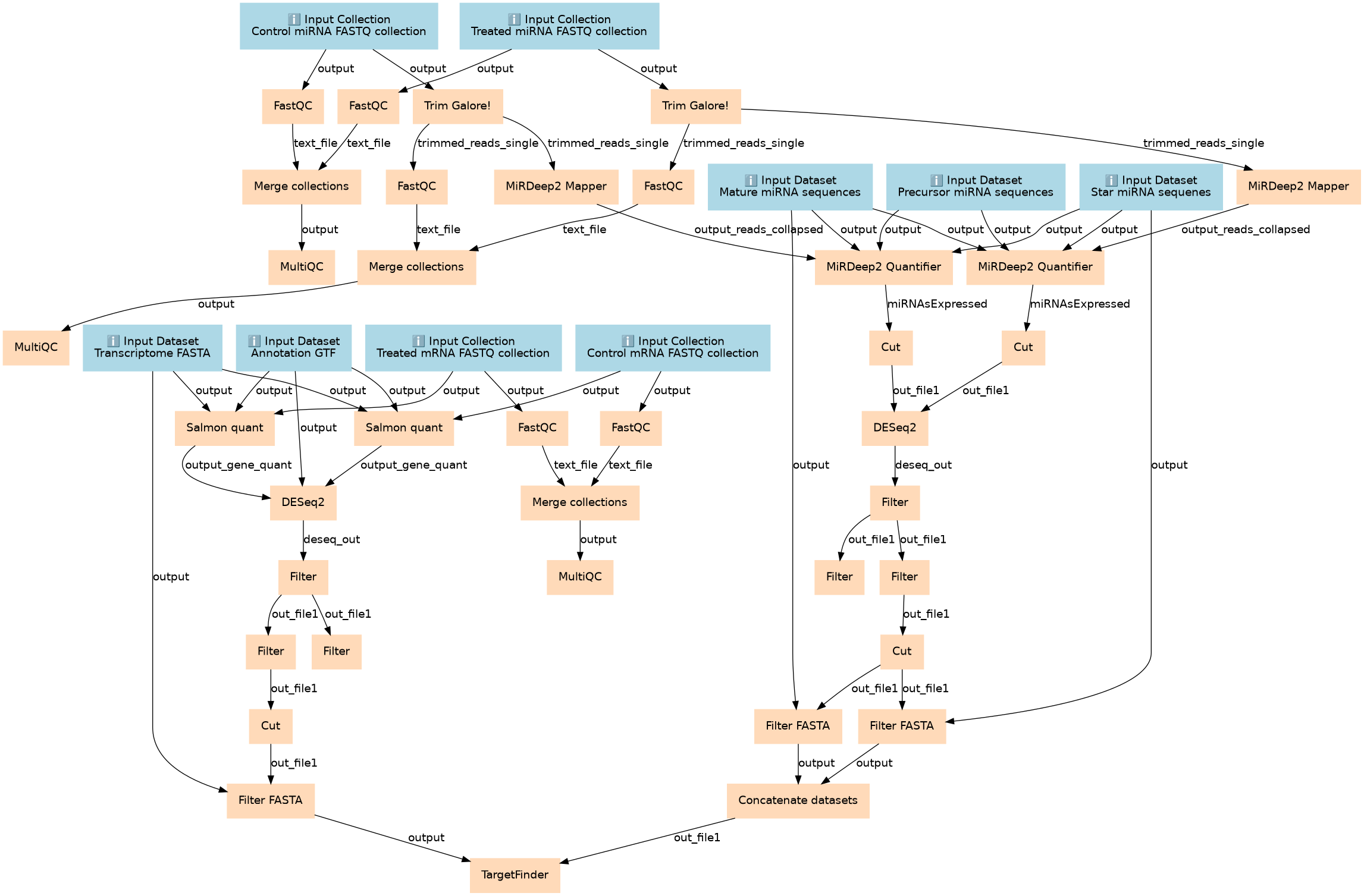
Identify upregulated miRNAS and analyze potential targets in downregulated genes.
Associated Tutorial
This workflows is part of the tutorial Whole transcriptome analysis of Arabidopsis thaliana, available in the GTN
Thanks to...
Tutorial Author(s): Cristóbal Gallardo, Pavankumar Videm, Beatriz Serrano-Solano
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Annotation GTF | #main/Annotation GTF | n/a |
|
| Control mRNA FASTQ collection | #main/Control mRNA FASTQ collection | n/a |
|
| Control miRNA FASTQ collection | #main/Control miRNA FASTQ collection | n/a |
|
| Mature miRNA sequences | #main/Mature miRNA sequences | n/a |
|
| Precursor miRNA sequences | #main/Precursor miRNA sequences | n/a |
|
| Star miRNA sequenes | #main/Star miRNA sequenes | n/a |
|
| Transcriptome FASTA | #main/Transcriptome FASTA | n/a |
|
| Treated mRNA FASTQ collection | #main/Treated mRNA FASTQ collection | n/a |
|
| Treated miRNA FASTQ collection | #main/Treated miRNA FASTQ collection | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 9 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 10 | Trim Galore! | toolshed.g2.bx.psu.edu/repos/bgruening/trim_galore/trim_galore/0.4.3.1 |
| 11 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 12 | Trim Galore! | toolshed.g2.bx.psu.edu/repos/bgruening/trim_galore/trim_galore/0.4.3.1 |
| 13 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 14 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 15 | Salmon quant | toolshed.g2.bx.psu.edu/repos/bgruening/salmon/salmon/0.14.1.2+galaxy1 |
| 16 | Salmon quant | toolshed.g2.bx.psu.edu/repos/bgruening/salmon/salmon/0.14.1.2+galaxy1 |
| 17 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 18 | MiRDeep2 Mapper | toolshed.g2.bx.psu.edu/repos/rnateam/mirdeep2_mapper/rbc_mirdeep2_mapper/2.0.0 |
| 19 | Merge collections | __MERGE_COLLECTION__ |
| 20 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.72+galaxy1 |
| 21 | MiRDeep2 Mapper | toolshed.g2.bx.psu.edu/repos/rnateam/mirdeep2_mapper/rbc_mirdeep2_mapper/2.0.0 |
| 22 | Merge collections | __MERGE_COLLECTION__ |
| 23 | DESeq2 | toolshed.g2.bx.psu.edu/repos/iuc/deseq2/deseq2/2.11.40.6+galaxy1 |
| 24 | MiRDeep2 Quantifier | toolshed.g2.bx.psu.edu/repos/rnateam/mirdeep2_quantifier/rbc_mirdeep2_quantifier/2.0.0 |
| 25 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.7 |
| 26 | Merge collections | __MERGE_COLLECTION__ |
| 27 | MiRDeep2 Quantifier | toolshed.g2.bx.psu.edu/repos/rnateam/mirdeep2_quantifier/rbc_mirdeep2_quantifier/2.0.0 |
| 28 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.7 |
| 29 | Filter | Filter1 |
| 30 | Cut | Cut1 |
| 31 | MultiQC | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.7 |
| 32 | Cut | Cut1 |
| 33 | Filter | Filter1 |
| 34 | Filter | Filter1 |
| 35 | DESeq2 | toolshed.g2.bx.psu.edu/repos/iuc/deseq2/deseq2/2.11.40.6+galaxy1 |
| 36 | Cut | Cut1 |
| 37 | Filter | Filter1 |
| 38 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.1 |
| 39 | Filter | Filter1 |
| 40 | Filter | Filter1 |
| 41 | Cut | Cut1 |
| 42 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.1 |
| 43 | Filter FASTA | toolshed.g2.bx.psu.edu/repos/galaxyp/filter_by_fasta_ids/filter_by_fasta_ids/2.1 |
| 44 | Concatenate datasets | cat1 |
| 45 | TargetFinder | toolshed.g2.bx.psu.edu/repos/rnateam/targetfinder/targetfinder/1.7.0+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 289 Downloads: 36 Runs: 0
Created: 2nd Jun 2025 at 11:06
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master