Workflow Type: Galaxy
Nucleoli segmentation and feature extraction using CellProfiler
Associated Tutorial
This workflows is part of the tutorial Nucleoli segmentation and feature extraction using CellProfiler, available in the GTN
Thanks to...
Tutorial Author(s): Beatriz Serrano-Solano, Jean-Karim Hériché
Tutorial Contributor(s): Helena Rasche, Saskia Hiltemann, Björn Grüning, Beatriz Serrano-Solano, Anne Fouilloux
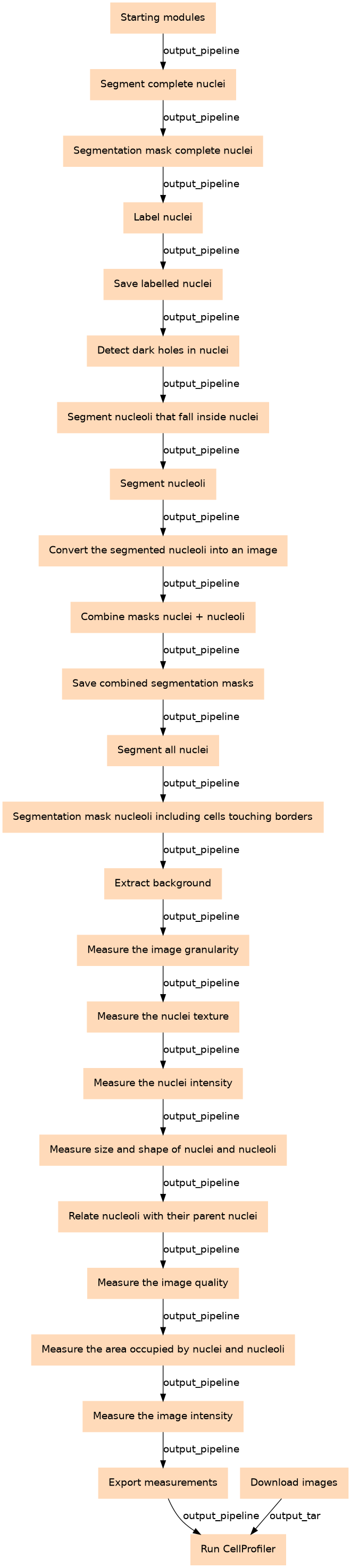
Steps
| ID | Name | Description |
|---|---|---|
| 0 | Starting modules | Detail the metadata associated to the images that will be processed toolshed.g2.bx.psu.edu/repos/bgruening/cp_common/cp_common/3.1.9 |
| 1 | Download images | toolshed.g2.bx.psu.edu/repos/iuc/idr_download_by_ids/idr_download_by_ids/0.41 |
| 2 | Segment complete nuclei | Incomplete nuclei that are touching the borders of the image are ignored and therefore, not segmented. It doesn't include the nuclei outside the diameter range either. toolshed.g2.bx.psu.edu/repos/bgruening/cp_identify_primary_objects/cp_identify_primary_objects/3.1.9 |
| 3 | Segmentation mask complete nuclei | toolshed.g2.bx.psu.edu/repos/bgruening/cp_convert_objects_to_image/cp_convert_objects_to_image/3.1.9 |
| 4 | Label nuclei | toolshed.g2.bx.psu.edu/repos/bgruening/cp_display_data_on_image/cp_display_data_on_image/3.1.9 |
| 5 | Save labelled nuclei | toolshed.g2.bx.psu.edu/repos/bgruening/cp_save_images/cp_save_images/3.1.9 |
| 6 | Detect dark holes in nuclei | toolshed.g2.bx.psu.edu/repos/bgruening/cp_enhance_or_suppress_features/cp_enhance_or_suppress_features/3.1.9 |
| 7 | Segment nucleoli that fall inside nuclei | The dark holes enhanced in the previous step are segmented only if they are inside a nuclei toolshed.g2.bx.psu.edu/repos/bgruening/cp_mask_image/cp_mask_image/3.1.9 |
| 8 | Segment nucleoli | The segmentation only affects those inside the nuclei toolshed.g2.bx.psu.edu/repos/bgruening/cp_identify_primary_objects/cp_identify_primary_objects/3.1.9 |
| 9 | Convert the segmented nucleoli into an image | toolshed.g2.bx.psu.edu/repos/bgruening/cp_convert_objects_to_image/cp_convert_objects_to_image/3.1.9 |
| 10 | Combine masks (nuclei + nucleoli) | toolshed.g2.bx.psu.edu/repos/bgruening/cp_gray_to_color/cp_gray_to_color/3.1.9 |
| 11 | Save combined segmentation masks | toolshed.g2.bx.psu.edu/repos/bgruening/cp_save_images/cp_save_images/3.1.9 |
| 12 | Segment all nuclei | Incomplete nuclei that are touching the borders of the image are segmented, also the nuclei outside the diameter range. toolshed.g2.bx.psu.edu/repos/bgruening/cp_identify_primary_objects/cp_identify_primary_objects/3.1.9 |
| 13 | Segmentation mask nucleoli including cells touching borders | toolshed.g2.bx.psu.edu/repos/bgruening/cp_convert_objects_to_image/cp_convert_objects_to_image/3.1.9 |
| 14 | Extract background | toolshed.g2.bx.psu.edu/repos/bgruening/cp_image_math/cp_image_math/0.1.0 |
| 15 | Measure the image granularity | This step measures the granularity of the original image toolshed.g2.bx.psu.edu/repos/bgruening/cp_measure_granularity/cp_measure_granularity/3.1.9 |
| 16 | Measure the nuclei texture | This step measures the texture of the original image and nuclei in it toolshed.g2.bx.psu.edu/repos/bgruening/cp_measure_texture/cp_measure_texture/3.1.9 |
| 17 | Measure the nuclei intensity | This step measures the intensity of the original image and the nuclei toolshed.g2.bx.psu.edu/repos/bgruening/cp_measure_object_intensity/cp_measure_object_intensity/3.1.9 |
| 18 | Measure size and shape of nuclei and nucleoli | toolshed.g2.bx.psu.edu/repos/bgruening/cp_measure_object_size_shape/cp_measure_object_size_shape/3.1.9 |
| 19 | Relate nucleoli with their parent nuclei | toolshed.g2.bx.psu.edu/repos/bgruening/cp_relate_objects/cp_relate_objects/3.1.9 |
| 20 | Measure the image quality | toolshed.g2.bx.psu.edu/repos/bgruening/cp_measure_image_quality/cp_measure_image_quality/3.1.9 |
| 21 | Measure the area occupied by nuclei and nucleoli | toolshed.g2.bx.psu.edu/repos/bgruening/cp_measure_image_area_occupied/cp_measure_image_area_occupied/3.1.9 |
| 22 | Measure the image intensity | toolshed.g2.bx.psu.edu/repos/bgruening/cp_measure_image_intensity/cp_measure_image_intensity/3.1.9 |
| 23 | Export measurements | toolshed.g2.bx.psu.edu/repos/bgruening/cp_export_to_spreadsheet/cp_export_to_spreadsheet/3.1.9 |
| 24 | Run CellProfiler | toolshed.g2.bx.psu.edu/repos/bgruening/cp_cellprofiler/cp_cellprofiler/3.1.9 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_10 | #main/_anonymous_output_10 | n/a |
|
| _anonymous_output_11 | #main/_anonymous_output_11 | n/a |
|
| _anonymous_output_12 | #main/_anonymous_output_12 | n/a |
|
| _anonymous_output_13 | #main/_anonymous_output_13 | n/a |
|
| _anonymous_output_14 | #main/_anonymous_output_14 | n/a |
|
| _anonymous_output_15 | #main/_anonymous_output_15 | n/a |
|
| _anonymous_output_16 | #main/_anonymous_output_16 | n/a |
|
| _anonymous_output_17 | #main/_anonymous_output_17 | n/a |
|
| _anonymous_output_18 | #main/_anonymous_output_18 | n/a |
|
| _anonymous_output_19 | #main/_anonymous_output_19 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_20 | #main/_anonymous_output_20 | n/a |
|
| _anonymous_output_21 | #main/_anonymous_output_21 | n/a |
|
| _anonymous_output_22 | #main/_anonymous_output_22 | n/a |
|
| _anonymous_output_23 | #main/_anonymous_output_23 | n/a |
|
| _anonymous_output_24 | #main/_anonymous_output_24 | n/a |
|
| _anonymous_output_25 | #main/_anonymous_output_25 | n/a |
|
| _anonymous_output_26 | #main/_anonymous_output_26 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
| _anonymous_output_5 | #main/_anonymous_output_5 | n/a |
|
| _anonymous_output_6 | #main/_anonymous_output_6 | n/a |
|
| _anonymous_output_7 | #main/_anonymous_output_7 | n/a |
|
| _anonymous_output_8 | #main/_anonymous_output_8 | n/a |
|
| _anonymous_output_9 | #main/_anonymous_output_9 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
Activity
Views: 237 Downloads: 31 Runs: 0
Created: 2nd Jun 2025 at 10:56
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master