Workflow Type: Galaxy
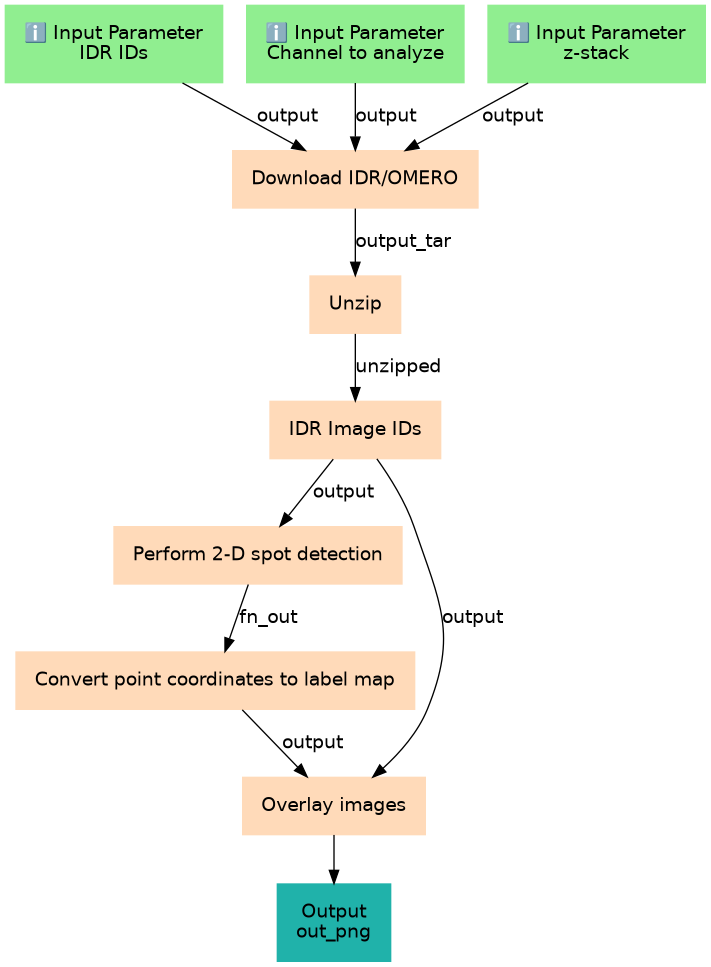
The present workflow allows to perform 2D spots/blobs detection on specific channel and stack of an image. Images will be directly fetched from IDR by including the IDs. A pre-processing steps with histogram normalization is performed before the blobs detection.
Associated Tutorial
This workflows is part of the tutorial Quantification of single-molecule RNA fluorescence in situ hybridization (smFISH) in yeast cell lines, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): Riccardo
Tutorial Author(s): Riccardo Massei
Tutorial Contributor(s): Leonid Kostrykin, Riccardo Massei, Björn Grüning
Funder(s): Deutsche Forschungsgemeinschaft
Grants(s): NFDI4Bioimage
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Channel to analyze | Channel to analyze | Name of the image channel to analyze |
|
| IDR IDs | IDR IDs | A list of images IDs from IDR |
|
| z-stack | z-stack | Select a z-stack to analyze |
|
Steps
| ID | Name | Description |
|---|---|---|
| 3 | Download IDR/OMERO | toolshed.g2.bx.psu.edu/repos/iuc/idr_download_by_ids/idr_download_by_ids/0.45 |
| 4 | Unzip | toolshed.g2.bx.psu.edu/repos/imgteam/unzip/unzip/6.0+galaxy0 |
| 5 | IDR Image IDs | List of images from IDR to analyzed toolshed.g2.bx.psu.edu/repos/imgteam/2d_histogram_equalization/ip_histogram_equalization/0.18.1+galaxy0 |
| 6 | Perform 2-D spot detection | toolshed.g2.bx.psu.edu/repos/imgteam/spot_detection_2d/ip_spot_detection_2d/0.1+galaxy0 |
| 7 | Convert point coordinates to label map | toolshed.g2.bx.psu.edu/repos/imgteam/points2labelimage/ip_points_to_label/0.4+galaxy0 |
| 8 | Overlay images | toolshed.g2.bx.psu.edu/repos/imgteam/overlay_images/ip_overlay_images/0.0.4+galaxy4 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| out_png | out_png | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
License
Activity
Views: 302 Downloads: 33 Runs: 0
Created: 2nd Jun 2025 at 10:57
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master