Workflow Type: Galaxy
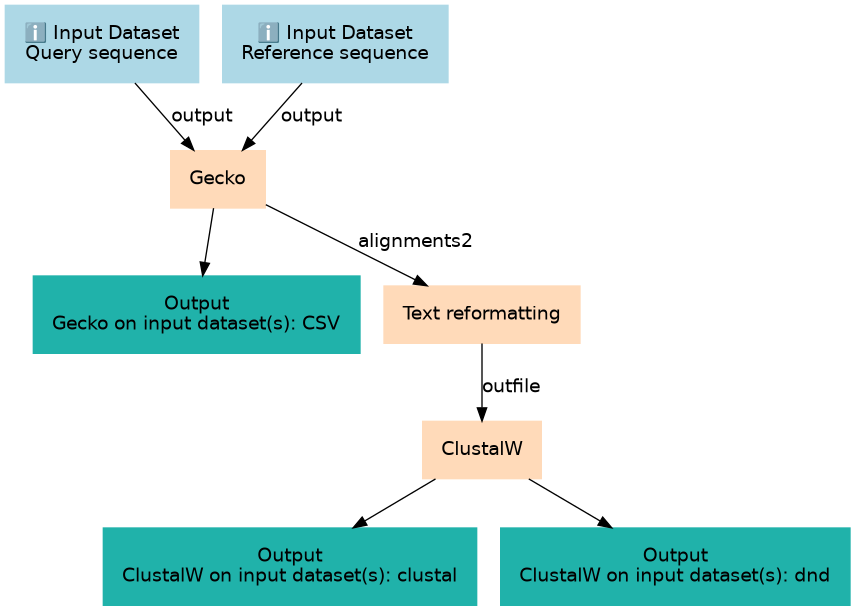
A workflow designed to compare two sequences using GECKO, extract repeats and perform multiple sequence alignment
Associated Tutorial
This workflows is part of the tutorial From small to large-scale genome comparison, available in the GTN
Thanks to...
Tutorial Author(s): Esteban Perez-Wohlfeil
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Query sequence | #main/Query sequence | n/a |
|
| Reference sequence | #main/Reference sequence | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Gecko | toolshed.g2.bx.psu.edu/repos/iuc/gecko/gecko/1.2 |
| 3 | Text reformatting | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_awk_tool/1.1.2 |
| 4 | ClustalW | toolshed.g2.bx.psu.edu/repos/devteam/clustalw/clustalw/2.1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| ClustalW on input dataset(s): clustal | #main/ClustalW on input dataset(s): clustal | n/a |
|
| ClustalW on input dataset(s): dnd | #main/ClustalW on input dataset(s): dnd | n/a |
|
| Gecko on input dataset(s): CSV | #main/Gecko on input dataset(s): CSV | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 254 Downloads: 35 Runs: 0
Created: 2nd Jun 2025 at 10:57
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master