Workflow Type: Galaxy
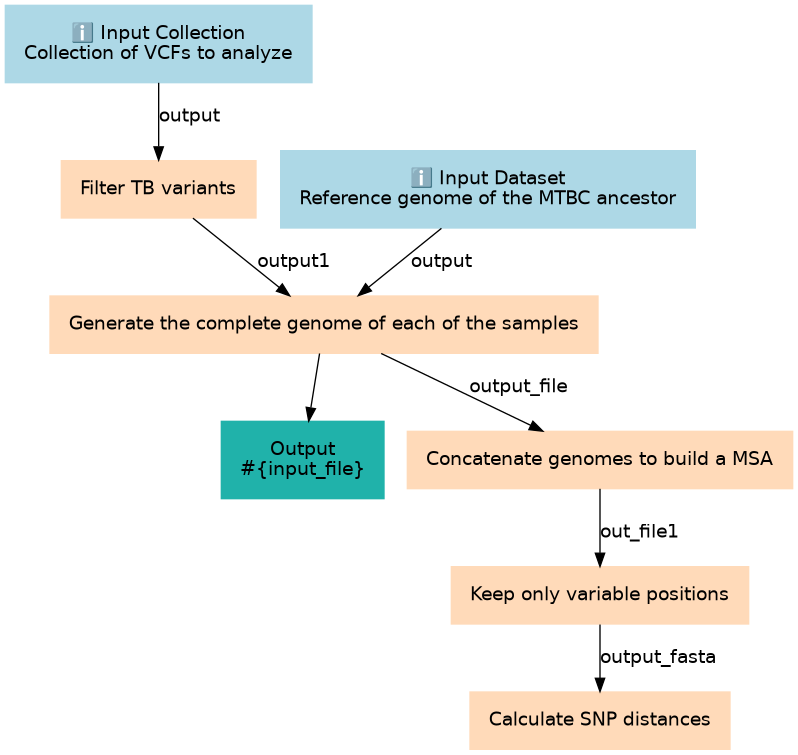
Given a set of VCF files and the reference genome used to do the mapping and SNP calling, create a multifasta file containing the genomes of all samples and calculate the matrix of pairwise SNP distances
Associated Tutorial
This workflows is part of the tutorial Identifying tuberculosis transmission links: from SNPs to transmission clusters, available in the GTN
Thanks to...
Tutorial Author(s): Galo A. Goig, Daniela Brites, Christoph Stritt
Tutorial Contributor(s): Wolfgang Maier, Saskia Hiltemann, Helena Rasche, Galo A. Goig, Björn Grüning, Peter van Heusden, Christoph Stritt, Lucille Delisle
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Collection of VCFs to analyze | #main/Collection of VCFs to analyze | n/a |
|
| Reference genome of the MTBC ancestor | #main/Reference genome of the MTBC ancestor | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Filter TB variants | We will ensure at this step that variants to build the MSA are fixed variants and that we low-confidence filter repetitive regions of the MTB genome toolshed.g2.bx.psu.edu/repos/iuc/tb_variant_filter/tb_variant_filter/0.1.3+galaxy0 |
| 3 | Generate the complete genome of each of the samples | The complete genome of each of the samples is generated by inserting the SNPs defined in the respective VCF in the reference genome that was used for mapping and SNP calling toolshed.g2.bx.psu.edu/repos/iuc/bcftools_consensus/bcftools_consensus/1.9+galaxy2 |
| 4 | Concatenate genomes to build a MSA | All genomes are concatenated in a single multifasta file. Because all o them have the same length, this may be seen as a multiple sequence alignment. toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cat/0.1.1 |
| 5 | Keep only variable positions | Discard invariant positions from the MSA to simplify the file so only contains positions with at least one SNP in at least one strain. toolshed.g2.bx.psu.edu/repos/iuc/snp_sites/snp_sites/2.5.1+galaxy0 |
| 6 | Calculate SNP distances | From the MSA. Calculate pairwise SNP distances between samples. toolshed.g2.bx.psu.edu/repos/iuc/snp_dists/snp_dists/0.6.3+galaxy0 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| {input_file} | #main/{input_file} | n/a |
|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
| _anonymous_output_3 | #main/_anonymous_output_3 | n/a |
|
| _anonymous_output_4 | #main/_anonymous_output_4 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tools
Activity
Views: 307 Downloads: 32 Runs: 0
Created: 2nd Jun 2025 at 10:59
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master